Next-Generation Sequencing-Based Genomic Profiling of Children with Acute Myeloid Leukemia
- PMID: 37088137
- PMCID: PMC10435843
- DOI: 10.1016/j.jmoldx.2023.04.004
Next-Generation Sequencing-Based Genomic Profiling of Children with Acute Myeloid Leukemia
Abstract
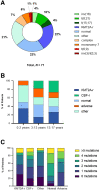
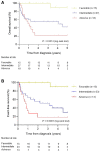
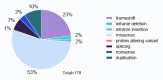
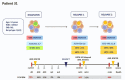
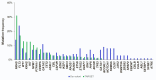
Pediatric acute myeloid leukemia (AML) represents a major cause of childhood leukemic mortality, with only a limited number of studies investigating the molecular landscape of the disease. Here, we present an integrative analysis of cytogenetic and molecular profiles of 75 patients with pediatric AML from a multicentric, real-world patient cohort treated according to AML Berlin-Frankfurt-Münster protocols. Targeted next-generation sequencing of 54 genes revealed 17 genes that were recurrently mutated in >5% of patients. Considerable differences were observed in the mutational profiles compared with previous studies, as BCORL1, CUX1, KDM6A, PHF6, and STAG2 mutations were detected at a higher frequency than previously reported, whereas KIT, NRAS, and KRAS were less frequently mutated. Our study identified novel recurrent mutations at diagnosis in the BCORL1 gene in 9% of the patients. Tumor suppressor gene (PHF6, TP53, and WT1) mutations were found to be associated with induction failure and shorter event-free survival, suggesting important roles of these alterations in resistance to therapy and disease progression. Comparison of the mutational landscape at diagnosis and relapse revealed an enrichment of mutations in tumor suppressor genes (16.2% versus 44.4%) and transcription factors (35.1% versus 55.6%) at relapse. Our findings shed further light on the heterogeneity of pediatric AML and identify previously unappreciated alterations that may lead to improved molecular characterization and risk stratification of pediatric AML.
Copyright © 2023 Association for Molecular Pathology and American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures











References
-
- Miller K.D., Nogueira L., Devasia T., Mariotto A.B., Yabroff K.R., Jemal A., Kramer J., Siegel R.L. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin. 2022;72:409–436. - PubMed
-
- Zwaan C.M., Kolb E.A., Reinhardt D., Abrahamsson J., Adachi S., Aplenc R., De Bont E.S.J.M., De Moerloose B., Dworzak M., Gibson B.E.S., Hasle H., Leverger G., Locatelli F., Ragu C., Ribeiro R.C., Rizzari C., Rubnitz J.E., Smith O.P., Sung L., Tomizawa D., van den Heuvel-Eibrink M.M., Creutzig U., Kaspers G.J.L. Collaborative efforts driving progress in pediatric acute myeloid leukemia. J Clin Oncol. 2015;33:2949–2962. - PMC - PubMed
-
- Rasche M., Zimmermann M., Borschel L., Bourquin J.-P., Dworzak M., Klingebiel T., Lehrnbecher T., Creutzig U., Klusmann J.-H., Reinhardt D. Successes and challenges in the treatment of pediatric acute myeloid leukemia: a retrospective analysis of the AML-BFM trials from 1987 to 2012. Leukemia. 2018;32:2167–2177. - PMC - PubMed
-
- Aplenc R., Meshinchi S., Sung L., Alonzo T., Choi J., Fisher B., Gerbing R., Hirsch B., Horton T., Kahwash S., Levine J., Loken M., Brodersen L., Pollard J., Raimondi S., Kolb E.A., Gamis A. Bortezomib with standard chemotherapy for children with acute myeloid leukemia does not improve treatment outcomes: a report from the Children’s Oncology Group. Haematologica. 2020;105:1879–1886. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

