In-silico target prediction by ensemble chemogenomic model based on multi-scale information of chemical structures and protein sequences
- PMID: 37088813
- PMCID: PMC10123967
- DOI: 10.1186/s13321-023-00720-0
In-silico target prediction by ensemble chemogenomic model based on multi-scale information of chemical structures and protein sequences
Abstract
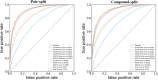
Identification and validation of bioactive small-molecule targets is a significant challenge in drug discovery. In recent years, various in-silico approaches have been proposed to expedite time- and resource-consuming experiments for target detection. Herein, we developed several chemogenomic models for target prediction based on multi-scale information of chemical structures and protein sequences. By combining the information of a compound with multiple protein targets together and putting these compound-target pairs into a well-established model, the scores to indicate whether there are interactions between compounds and targets can be derived, and thus a target prediction task can be completed by sorting the outputted scores. To improve the prediction performance, we constructed several chemogenomic models using multi-scale information of chemical structures and protein sequences, and the ensemble model with the best performance was used as our final model. The model was validated by various strategies and external datasets and the promising target prediction capability of the model, i.e., the fraction of known targets identified in the top-k (1 to 10) list of the potential target candidates suggested by the model, was confirmed. Compared with multiple state-of-art target prediction methods, our model showed equivalent or better predictive ability in terms of the top-k predictions. It is expected that our method can be utilized as a powerful computational tool to narrow down the potential targets for experimental testing.
Keywords: Chemogenomic; Ensemble model; Target prediction; XGBoost.
© 2023. The Author(s).
Conflict of interest statement
The authors declare neither competing financial interests nor non-financial competing interests.
Figures




Similar articles
-
Chemogenomic Approaches for Revealing Drug Target Interactions in Drug Discovery.Curr Genomics. 2021 Dec 30;22(5):328-338. doi: 10.2174/1389202922666210920125800. Curr Genomics. 2021. PMID: 35283667 Free PMC article. Review.
-
Benchmarking a Wide Range of Chemical Descriptors for Drug-Target Interaction Prediction Using a Chemogenomic Approach.Mol Inform. 2014 Dec;33(11-12):719-31. doi: 10.1002/minf.201400066. Epub 2014 Nov 24. Mol Inform. 2014. PMID: 27485418 Review.
-
TargetHunter: an in silico target identification tool for predicting therapeutic potential of small organic molecules based on chemogenomic database.AAPS J. 2013 Apr;15(2):395-406. doi: 10.1208/s12248-012-9449-z. Epub 2013 Jan 5. AAPS J. 2013. PMID: 23292636 Free PMC article.
-
Drug-target interaction prediction using ensemble learning and dimensionality reduction.Methods. 2017 Oct 1;129:81-88. doi: 10.1016/j.ymeth.2017.05.016. Epub 2017 May 24. Methods. 2017. PMID: 28549952
-
A comprehensive review of feature based methods for drug target interaction prediction.J Biomed Inform. 2019 May;93:103159. doi: 10.1016/j.jbi.2019.103159. Epub 2019 Mar 27. J Biomed Inform. 2019. PMID: 30926470 Review.
Cited by
-
Enhancing the confidence of potential targets enriched by similarity-centric models: the crucial role of the similarity threshold.Front Pharmacol. 2025 Aug 8;16:1574540. doi: 10.3389/fphar.2025.1574540. eCollection 2025. Front Pharmacol. 2025. PMID: 40860884 Free PMC article.
-
Synthesis of Pyrrolo[3,4-b]pyridin-5-ones via Ugi-Zhu Reaction and In Vitro-In Silico Studies against Breast Carcinoma.Pharmaceuticals (Basel). 2023 Nov 6;16(11):1562. doi: 10.3390/ph16111562. Pharmaceuticals (Basel). 2023. PMID: 38004428 Free PMC article.
-
Bioengineering approaches to trained immunity: Physiologic targets and therapeutic strategies.Elife. 2025 Jul 23;14:e106339. doi: 10.7554/eLife.106339. Elife. 2025. PMID: 40699210 Free PMC article. Review.
-
Computational Insights into the Interaction of Pinostrobin with Bcl-2 Family Proteins: A Molecular Docking Analysis.Asian Pac J Cancer Prev. 2024 Feb 1;25(2):507-512. doi: 10.31557/APJCP.2024.25.2.507. Asian Pac J Cancer Prev. 2024. PMID: 38415536 Free PMC article.
-
CACTI: an in silico chemical analysis tool through the integration of chemogenomic data and clustering analysis.J Cheminform. 2024 Jul 24;16(1):84. doi: 10.1186/s13321-024-00885-2. J Cheminform. 2024. PMID: 39049122 Free PMC article.
References
Grants and funding
- SDF19 0402 P02/HKBU Strategic Development Fund project
- 2021YFF1201400/National Key Research and Development Program of China
- 22173118/National Natural Science Foundation of China
- 2021JJ10068/Hunan Provincial Science Fund for Distinguished Young Scholars
- TC210804V/The Project of Intelligent Management Software for Multimodal Medical Big Data for New Generation Information Technology, Ministry of Industry and Information Technology of People's Republic of China
LinkOut - more resources
Full Text Sources

