This is a preprint.
Utilizing river and wastewater as a SARS-CoV-2 surveillance tool to predict trends and identify variants of concern in settings with limited formal sewage systems
- PMID: 37090541
- PMCID: PMC10120776
- DOI: 10.21203/rs.3.rs-2801767/v1
Utilizing river and wastewater as a SARS-CoV-2 surveillance tool to predict trends and identify variants of concern in settings with limited formal sewage systems
Update in
-
Utilizing river and wastewater as a SARS-CoV-2 surveillance tool in settings with limited formal sewage systems.Nat Commun. 2023 Nov 30;14(1):7883. doi: 10.1038/s41467-023-43047-y. Nat Commun. 2023. PMID: 38036496 Free PMC article.
Abstract
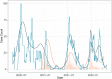
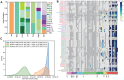
The COVID-19 pandemic continues to impact health systems globally and robust surveillance is critical for pandemic control, however not all countries can sustain community surveillance programs. Wastewater surveillance has proven valuable in high-income settings, but little is known about how river and informal sewage in low-income countries can be used for environmental surveillance of SARS-CoV-2. In Malawi, a country with limited community-based COVID-19 testing capacity, we explored the utility of rivers and wastewater for SARS-CoV-2 surveillance. From May 2020 - January 2022, we collected water from up to 112 river or informal sewage sites/month, detecting SARS-CoV-2 in 8.3% of samples. Peak SARS-CoV-2 detection in water samples predated peaks in clinical cases. Sequencing of water samples identified the Beta, Delta, and Omicron variants, with Delta and Omicron detected well in advance of detection in patients. Our work highlights wastewater can be used for detecting emerging waves, identifying variants of concern and function as an early warning system in settings with no formal sewage systems.
Figures

References
-
- Chibwana M. G. et al. High SARS-CoV-2 seroprevalence in health care workers but relatively low numbers of deaths in urban Malawi. medRxiv, doi: 10.1101/2020.07.30.20164970 (2020). - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

