This is a preprint.
Widespread CRISPR repeat-like RNA regulatory elements in CRISPR-Cas systems
- PMID: 37090614
- PMCID: PMC10120712
- DOI: 10.1101/2023.03.03.530964
Widespread CRISPR repeat-like RNA regulatory elements in CRISPR-Cas systems
Update in
-
Widespread CRISPR-derived RNA regulatory elements in CRISPR-Cas systems.Nucleic Acids Res. 2023 Aug 25;51(15):8150-8168. doi: 10.1093/nar/gkad495. Nucleic Acids Res. 2023. PMID: 37283088 Free PMC article.
Abstract
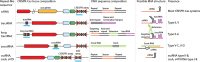
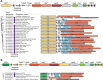
CRISPR- cas loci typically contain CRISPR arrays with unique spacers separating direct repeats. Spacers along with portions of adjacent repeats are transcribed and processed into CRISPR(cr) RNAs that target complementary sequences (protospacers) in mobile genetic elements, resulting in cleavage of the target DNA or RNA. Additional, standalone repeats in some CRISPR- cas loci produce distinct cr-like RNAs implicated in regulatory or other functions. We developed a computational pipeline to systematically predict crRNA-like elements by scanning for standalone repeat sequences that are conserved in closely related CRISPR- cas loci. Numerous crRNA-like elements were detected in diverse CRISPR-Cas systems, mostly, of type I, but also subtype V-A. Standalone repeats often form mini-arrays containing two repeat-like sequence separated by a spacer that is partially complementary to promoter regions of cas genes, in particular cas8 , or cargo genes located within CRISPR-Cas loci, such as toxins-antitoxins. We show experimentally that a mini-array from a type I-F1 CRISPR-Cas system functions as a regulatory guide. We also identified mini-arrays in bacteriophages that could abrogate CRISPR immunity by inhibiting effector expression. Thus, recruitment of CRISPR effectors for regulatory functions via spacers with partial complementarity to the target is a common feature of diverse CRISPR-Cas systems.
Conflict of interest statement
Conflict statement
The authors report no conflict of interest.
Figures







Similar articles
-
Widespread CRISPR-derived RNA regulatory elements in CRISPR-Cas systems.Nucleic Acids Res. 2023 Aug 25;51(15):8150-8168. doi: 10.1093/nar/gkad495. Nucleic Acids Res. 2023. PMID: 37283088 Free PMC article.
-
The CRISPR Spacer Space Is Dominated by Sequences from Species-Specific Mobilomes.mBio. 2017 Sep 19;8(5):e01397-17. doi: 10.1128/mBio.01397-17. mBio. 2017. PMID: 28928211 Free PMC article.
-
Anti-cas spacers in orphan CRISPR4 arrays prevent uptake of active CRISPR-Cas I-F systems.Nat Microbiol. 2016 Jun 6;1(8):16081. doi: 10.1038/nmicrobiol.2016.81. Nat Microbiol. 2016. PMID: 27573106
-
Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity.FEMS Microbiol Rev. 2015 May;39(3):428-41. doi: 10.1093/femsre/fuv023. Epub 2015 May 19. FEMS Microbiol Rev. 2015. PMID: 25994611 Free PMC article. Review.
-
Analysis of direct repeats and spacers of CRISPR/Cas systems type I-F in Brazilian clinical strains of Pseudomonas aeruginosa.Mol Genet Genomics. 2019 Oct;294(5):1095-1105. doi: 10.1007/s00438-019-01575-7. Epub 2019 May 16. Mol Genet Genomics. 2019. PMID: 31098740 Review.
References
-
- Mohanraju P., Makarova K.S., Zetsche B., Zhang F., Koonin E.V. and van der Oost J. (2016) Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science, 353, aad5147. - PubMed
-
- Barrangou R. and Horvath P. (2017) A decade of discovery: CRISPR functions and applications. Nat Microbiol, 2, 17092. - PubMed
-
- Nussenzweig P.M. and Marraffini L.A. (2020) Molecular Mechanisms of CRISPR-Cas Immunity in Bacteria. Annu Rev Genet, 54, 93–120. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
