This is a preprint.
Loss of MC1R signaling implicates TBX3 in pheomelanogenesis and melanoma predisposition
- PMID: 37090624
- PMCID: PMC10120706
- DOI: 10.1101/2023.03.10.532018
Loss of MC1R signaling implicates TBX3 in pheomelanogenesis and melanoma predisposition
Abstract
The human Red Hair Color (RHC) trait is caused by increased pheomelanin (red-yellow) and reduced eumelanin (black-brown) pigment in skin and hair due to diminished melanocortin 1 receptor (MC1R) function. In addition, individuals harboring the RHC trait are predisposed to melanoma development. While MC1R variants have been established as causative of RHC and are a well-defined risk factor for melanoma, it remains unclear mechanistically why decreased MC1R signaling alters pigmentation and increases melanoma susceptibility. Here, we use single-cell RNA-sequencing (scRNA-seq) of melanocytes isolated from RHC mouse models to reveal a Pheomelanin Gene Signature (PGS) comprising genes implicated in melanogenesis and oncogenic transformation. We show that TBX3, a well-known anti-senescence transcription factor implicated in melanoma progression, is part of the PGS and binds both E-box and T-box elements to regulate genes associated with melanogenesis and senescence bypass. Our results provide key insights into mechanisms by which MC1R signaling regulates pigmentation and how individuals with the RHC phenotype are predisposed to melanoma.
Keywords: TBX3; melanocortin 1 receptor (MC1R); melanocytes; melanoma; red hair color.
Conflict of interest statement
Competing Interest Statement: Authors have no competing interests to declare.
Figures

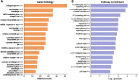

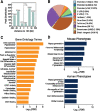
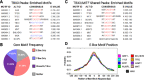
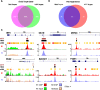
References
-
- Bennett D. C. & Medrano E. E. Molecular Regulation of Melanocyte Senescence. Pigm Cell Res 15, 242–250 (2002). - PubMed
-
- Rhodes A. R., Weinstock M. A., Fitzpatrick T. B., Mihm M. C. & Sober A. J. Risk Factors for Cutaneous Melanoma: A Practical Method of Recognizing Predisposed Individuals. Jama 258, 3146–3154 (1987). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
