This is a preprint.
The E592K variant of SF3B1 creates unique RNA missplicing and associates with high-risk MDS without ring sideroblasts
- PMID: 37090662
- PMCID: PMC10120771
- DOI: 10.21203/rs.3.rs-2802265/v1
The E592K variant of SF3B1 creates unique RNA missplicing and associates with high-risk MDS without ring sideroblasts
Update in
-
The E592K variant of SF3B1 creates unique RNA missplicing and associates with high-risk MDS without ring sideroblasts.Blood Adv. 2024 Aug 13;8(15):3961-3971. doi: 10.1182/bloodadvances.2023011260. Blood Adv. 2024. PMID: 38759096 Free PMC article.
Abstract
Among the most common genetic alterations in the myelodysplastic syndromes (MDS) are mutations in the spliceosome gene SF3B1. Such mutations induce specific RNA missplicing events, directly promote ring sideroblast (RS) formation, generally associate with more favorable prognosis, and serve as a predictive biomarker of response to luspatercept. However, not all SF3B1 mutations are the same, and here we report that the E592K variant of SF3B1 associates with high-risk disease features in MDS, including a lack of RS, increased myeloblasts, a distinct co-mutation pattern, and decreased survival. Moreover, in contrast to canonical SF3B1 mutations, E592K induces a unique RNA missplicing pattern, retains an interaction with the splicing factor SUGP1, and preserves normal RNA splicing of the sideroblastic anemia genes TMEM14C and ABCB7. These data expand our knowledge of the functional diversity of spliceosome mutations, and they suggest that patients with E592K should be approached differently from low-risk, luspatercept-responsive MDS patients with ring sideroblasts and canonical SF3B1 mutations.
Conflict of interest statement
Competing Interests The authors declare no competing interests.
Figures
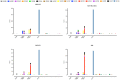
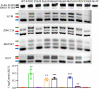
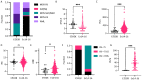



References
-
- Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3′ Splice Site Selection through Use of a Different Branch Point. Cell Reports 2015; 13: 1033–1045. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

