This is a preprint.
RAD52 prevents accumulation of -dependent replication gaps at perturbed replication forks in human cells
- PMID: 37090680
- PMCID: PMC10120653
- DOI: 10.1101/2023.04.12.536536
RAD52 prevents accumulation of -dependent replication gaps at perturbed replication forks in human cells
Abstract
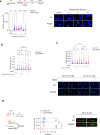
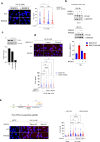
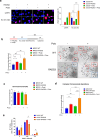
Replication gaps can arise as a consequence of perturbed DNA replication and their accumulation might undermine the stability of the genome. Loss of RAD52, a protein involved in the regulation of fork reversal, promotes accumulation of parental ssDNA gaps during replication perturbation. Here, we demonstrate that this is due to the engagement of downstream of the extensive degradation of perturbed replication forks after their reversal, and is not dependent on PrimPol. is hyper-recruited at parental ssDNA in the absence of RAD52, and this recruitment is dependent on fork reversal enzymes and RAD51. Of note, we report that the interaction between and RAD51 is stimulated by RAD52 inhibition, and -dependent gap accumulation requires nucleation of RAD51 suggesting that it occurs downstream strand invasion. Altogether, our data indicate that RAD51- -dependent repriming is essential to promote fork restart and limit DNA damage accumulation when RAD52 function is disabled.
Conflict of interest statement
CONFLICT OF INTEREST The authors declare to do not have any conflict of interest
Figures









Similar articles
-
Rad52 prevents excessive replication fork reversal and protects from nascent strand degradation.Nat Commun. 2019 Mar 29;10(1):1412. doi: 10.1038/s41467-019-09196-9. Nat Commun. 2019. PMID: 30926821 Free PMC article.
-
CHK1 phosphorylates PRIMPOL to promote replication stress tolerance.Sci Adv. 2022 Apr;8(13):eabm0314. doi: 10.1126/sciadv.abm0314. Epub 2022 Mar 30. Sci Adv. 2022. PMID: 35353580 Free PMC article.
-
WRNIP1 protects stalled forks from degradation and promotes fork restart after replication stress.EMBO J. 2016 Jul 1;35(13):1437-51. doi: 10.15252/embj.201593265. Epub 2016 May 30. EMBO J. 2016. PMID: 27242363 Free PMC article.
-
Non-Recombinogenic Functions of Rad51, BRCA2, and Rad52 in DNA Damage Tolerance.Genes (Basel). 2021 Sep 29;12(10):1550. doi: 10.3390/genes12101550. Genes (Basel). 2021. PMID: 34680945 Free PMC article. Review.
-
Physiological and Pathological Roles of RAD52 at DNA Replication Forks.Cancers (Basel). 2020 Feb 10;12(2):402. doi: 10.3390/cancers12020402. Cancers (Basel). 2020. PMID: 32050645 Free PMC article. Review.
Cited by
-
RAD52 and ERCC6L/PICH have a compensatory relationship for genome stability in mitosis.PLoS Genet. 2024 Nov 19;20(11):e1011479. doi: 10.1371/journal.pgen.1011479. eCollection 2024 Nov. PLoS Genet. 2024. PMID: 39561207 Free PMC article.
References
-
- Pasero P. & Vindigni A. Nucleases Acting at Stalled Forks: How to Reboot the Replication Program with a Few Shortcuts. Annu. Rev. Genet. 51, 477–499 (2017). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
