H3N2 influenza hemagglutination inhibition method qualification with data driven statistical methods for human clinical trials
- PMID: 37090729
- PMCID: PMC10117676
- DOI: 10.3389/fimmu.2023.1155880
H3N2 influenza hemagglutination inhibition method qualification with data driven statistical methods for human clinical trials
Abstract
Introduction: Hemagglutination inhibition (HAI) antibody titers to seasonal influenza strains are important surrogates for vaccine-elicited protection. However, HAI assays can be variable across labs, with low sensitivity across diverse viruses due to lack of standardization. Performing qualification of these assays on a strain specific level enables the precise and accurate quantification of HAI titers. Influenza A (H3N2) continues to be a predominant circulating subtype in most countries in Europe and North America since 1968 and is thus a focus of influenza vaccine research.
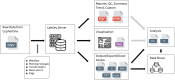
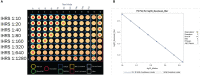
Methods: As a part of the National Institutes of Health (NIH)-funded Collaborative Influenza Vaccine Innovation Centers (CIVICs) program, we report on the identification of a robust assay design, rigorous statistical analysis, and complete qualification of an HAI assay using A/Texas/71/2017 as a representative H3N2 strain and guinea pig red blood cells and neuraminidase (NA) inhibitor oseltamivir to prevent NA-mediated agglutination.
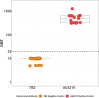
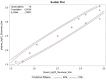
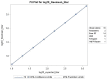
Results: This qualified HAI assay is precise (calculated by the geometric coefficient of variation (GCV)) for intermediate precision and intra-operator variability, accurate calculated by relative error, perfectly linear (slope of -1, R-Square 1), robust (<25% GCV) and depicts high specificity and sensitivity. This HAI method was successfully qualified for another H3N2 influenza strain A/Singapore/INFIMH-16-0019/2016, meeting all pre-specified acceptance criteria.
Discussion: These results demonstrate that HAI qualification and data generation for new influenza strains can be achieved efficiently with minimal extra testing and development. We report on a qualified and adaptable influenza serology method and analysis strategy to measure quantifiable HAI titers to define correlates of vaccine mediated protection in human clinical trials.
Keywords: antibody; clinical trials; data pipeline; hemagglutination inhibition (HAI); human; influenza; qualification; statistical analysis.
Copyright © 2023 Sawant, Gurley, Overman, Sharak, Mudrak, Oguin, Sempowski, Sarzotti-Kelsoe, Walter, Xie, Pasetti, Moody and Tomaras.
Conflict of interest statement
EBW has received funding support from Pfizer, Moderna, Sequiris, Najit Technologies Inc, and Clinetic for the conduct of clinical trials and clinical research. EBW has served as an advisor to Vaxcyte and consultant to ILiAD biotechnologies. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- World Health Organization . Influenza update n° 437 (2023). Available at: https://cdn.who.int/media/docs/default-source/influenza/influenza-update....
-
- Xie H, Li X, Gao J, Lin Z, Jing X, Plant E, et al. . Revisiting the 1976 "swine flu" vaccine clinical trials: cross-reactive hemagglutinin and neuraminidase antibodies and their role in protection against the 2009 H1N1 pandemic virus in mice. Clin Infect Dis (2011) 53(12):1179–87. doi: 10.1093/cid/cir693 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

