Novel Variant Identified in the Enhancer Region of Host Transcription Factor, BRN3A, is a Significant Risk Factor for HPV-Induced Uterine Cervix Cancer
- PMID: 37091039
- PMCID: PMC10116355
- DOI: 10.22088/IJMCM.BUMS.11.2.88
Novel Variant Identified in the Enhancer Region of Host Transcription Factor, BRN3A, is a Significant Risk Factor for HPV-Induced Uterine Cervix Cancer
Abstract
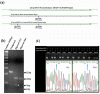
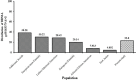
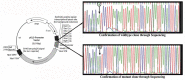
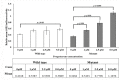
Among the HPV-mediated cervical cancers, cellular factor BRN3A has gained considerable attention due to its role in promoting an anti-apoptotic cellular environment and in facilitating epitheliotropic transformations of the host. The majority of previous studies looked at BRN3A's molecular characteristics; however, the possibility of genetic variations in BRN3A's auto-regulatory region in relation to cervical cancer risk has been underestimated until now. In a retrospective study in the Eastern UP population, India, we detected genetic variations in the cis-regulatory proximal enhancer region located around 5.6 kb upstream of transcription start site of BRN3A. Our analysis of PCR and DNA sequencing confirmed this novel SNP (BRN3A g.60163379A>G) within the auto-regulatory region of BRN3A. As compared to control subjects, cancer cases exhibited a 1.32-fold higher allele frequency (χ2 = 6.315, p = 0.012). In homozygous (GG) but not in heterozygous conditions, odds ratio (OR) analysis suggests a significant association of cancer risk with the SNP (OR = 2.60, p ≤ 0.004). We further confirmed using the functional analysis that this SNP increased the luciferase gene activity in HPV-positive cervical cancer SiHa cells that were exposed to progesterone. As a result of the association of polymorphisms in a non-coding region of an oncogene with increased cancer risks, we are suggesting that this genetic variation in non-coding region can be used in prediction, diagnosis, or predicting the progression of the disease.
Keywords: BRN3A; BRN3A proximal enhancer region; Cervical cancer; HPV; India; g. 6016 3379A >G.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Novel Brn3a cis-acting sequences mediate transcription of human trkA in neurons.J Neurochem. 2008 Apr;105(2):425-35. doi: 10.1111/j.1471-4159.2007.05139.x. Epub 2007 Dec 12. J Neurochem. 2008. PMID: 18086126
-
Genomic cloning and characterization of the nonoccupied allele corresponding to the integration site of human papillomavirus type 16 DNA in the cervical cancer cell line SiHa.Virology. 1996 Mar 1;217(1):33-41. doi: 10.1006/viro.1996.0090. Virology. 1996. PMID: 8599218
-
Polymorphism of the p53 codon 72 Arg/Pro and the risk of HPV type 16/18-associated cervical and oral cancer in India.Mol Cell Biochem. 2003 Oct;252(1-2):117-24. doi: 10.1023/a:1025546610920. Mol Cell Biochem. 2003. PMID: 14577584
-
On the etiology of anal squamous carcinoma.Dan Med Bull. 2002 Aug;49(3):194-209. Dan Med Bull. 2002. PMID: 12238281 Review.
-
The role of steroid contraceptive hormones in the pathogenesis of invasive cervical cancer: a review.Int J Gynecol Cancer. 2003 Mar-Apr;13(2):103-10. doi: 10.1046/j.1525-1438.2003.13030.x. Int J Gynecol Cancer. 2003. PMID: 12657108 Review.
References
-
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Bruni L, Albero G, Serrano B, et al. ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre), Human Papillomavirus and Related Diseases in India. Summary Report . 2019
-
- Srivastava S, Gupta S, Roy JK. High prevalence of oncogenic HPV-16 in cervical smears of asymptomatic women of eastern Uttar Pradesh, India: a population-based study. J Biosci. 2012;37:63–72. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
