Latent CSN-CRL complexes are crucial for curcumin-induced apoptosis and recruited during adipogenesis to lipid droplets via small GTPase RAB18
- PMID: 37091236
- PMCID: PMC10119602
- DOI: 10.1016/j.isci.2023.106468
Latent CSN-CRL complexes are crucial for curcumin-induced apoptosis and recruited during adipogenesis to lipid droplets via small GTPase RAB18
Abstract
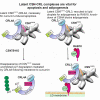
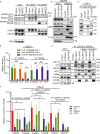
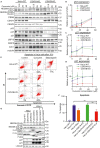
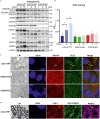
The COP9 signalosome (CSN) and cullin-RING ubiquitin ligases (CRLs) form latent CSN-CRL complexes detectable in cells. We demonstrate that the CSN variants CSNCSN7A and CSNCSN7B preferentially bind to CRL3 or CRL4A and CRL4B, respectively. Interestingly, the interacting protein ubiquitin-specific protease 15 exclusively binds to latent CSNCSN7A-CRL3, while p27KIP attaches to latent CSNCSN7B-CRL4A complex. Inhibition of deneddylation by CSN5i-3 or neddylation by MLN4924 do not impede the formation of latent complexes. Latent CSNCSN7A-CRL3 and latent CSNCSN7B-CRL4A/B particles are essential for specific cellular functions. We found that curcumin-induced cell death requires latent CSNCSN7B-CRL4A. Knockout of CSN7B in HeLa cells leads to resistance against curcumin. Remarkably, the small GTPase RAB18 recruits latent CSNCSN7A-CRL3 complex to lipid droplets (LDs), where CRL3 is activated by neddylation, an essential event for LD formation during adipogenesis. Knockdown of CSN7A or RAB18 or destabilization of latent complexes by cutting off CSN7A C-terminal 201-275 amino acids blocks adipogenesis.
Keywords: Cell biology; Cellular physiology; Functional aspects of cell biology.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Deng X.W., Dubiel W., Wei N., Hofmann K., Mundt K., Colicelli J., Kato J., Naumann M., Segal D., Seeger M., et al. Unified nomenclature for the COP9 signalosome and its subunits: an essential regulator of development. Trends Genet. 2000;16:203–289. - PubMed
-
- Hofmann K., Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem. Sci. 1998;23:204–205. - PubMed
-
- Lingaraju G.M., Bunker R.D., Cavadini S., Hess D., Hassiepen U., Renatus M., Fischer E.S., Thomä N.H. Crystal structure of the human COP9 signalosome. Nature. 2014;512:161–165. - PubMed
-
- Rockel B., Schmaler T., Huang X., Dubiel W. Electron microscopy and in vitro deneddylation reveal similar architectures and biochemistry of isolated human and Flag-mouse COP9 signalosome complexes. Biochem. Biophys. Res. Commun. 2014;450:991–997. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

