Neoadjuvant chemotherapy enhances anti-tumor immune response of tumor microenvironment in human esophageal squamous cell carcinoma
- PMID: 37091252
- PMCID: PMC10113784
- DOI: 10.1016/j.isci.2023.106480
Neoadjuvant chemotherapy enhances anti-tumor immune response of tumor microenvironment in human esophageal squamous cell carcinoma
Abstract
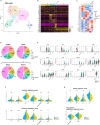
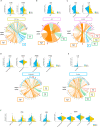
Although chemotherapy has been an essential treatment for cancer, the development of immune checkpoint blockade therapy was revolutionary, and a comprehensive understanding of the immunological tumor microenvironment (TME) has become crucial. Here, we investigated the impact of neoadjuvant chemotherapy (NAC) on immune cells in the TME of human esophageal squamous cell carcinoma using single cell RNA-sequencing. Analysis of 30 fresh samples revealed that CD8+/CD4+ T cells, dendritic cells (DCs), and macrophages in the TME of human esophageal squamous cell carcinoma showed higher levels of an anti-tumor immune response in the NAC(+) group than in the NAC(-) group. Furthermore, the immune cells of the NAC(+) group interacted with each other resulting in enhanced anti-tumor immune response via various cytokines, including IFNG in CD8+/CD4+ T cells, EBI3 in DCs, and NAMPT in macrophages. Our results suggest that NAC potentially enhances the anti-tumor immune response of immune cells in the TME.
Keywords: Cancer; Immunology; Transcriptomics.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Hiraki Y., Kimura Y., Imano M., Kato H., Iwama M., Shiraishi O., Yasuda A., Shinkai M., Makino T., Motoori M., et al. Controlling lymph node micrometastases by neoadjuvant chemotherapy affects the prognosis in advanced esophageal squamous cell carcinoma. Surg. Today. 2021;51:118–126. doi: 10.1007/s00595-020-02059-7. - DOI - PubMed
-
- Ando N., Kato H., Igaki H., Shinoda M., Ozawa S., Shimizu H., Nakamura T., Yabusaki H., Aoyama N., Kurita A., et al. A randomized trial comparing postoperative adjuvant chemotherapy with cisplatin and 5-fluorouracil versus preoperative chemotherapy for localized advanced squamous cell carcinoma of the thoracic esophagus (JCOG9907) Ann. Surg Oncol. 2012;19:68–74. doi: 10.1245/s10434-011-2049-9. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

