Taxonomic revision of Blumeria based on multi-gene DNA sequences, host preferences and morphology
- PMID: 37091321
- PMCID: PMC9157761
- DOI: 10.47371/mycosci.2020.12.003
Taxonomic revision of Blumeria based on multi-gene DNA sequences, host preferences and morphology
Abstract
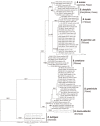
A taxonomic revision of the hitherto monotypic genus Blumeria was conducted incorporating multi-gene sequence analyses, host preference data and morphological criteria. The sequenced loci included rDNA ITS, partial chitin synthase gene (CHS1), as well as fragments of two unnamed orthologous genes (Bgt-1929, Bgt-4572). The combined evidence led to a reassessment and a new neotypification of B. graminiss. str. (emend.), and the description of seven additional species, viz. B. americana sp. nov. (mainly on hosts of the Triticeae), B. avenae sp. nov. (on Avena spp.), B. bromi-cathartici sp. nov. (on Bromus catharticus), B. bulbigera comb. nov. (on Bromus spp.), B. dactylidis sp. nov. (on Dactylis glomerata as the main host, but also on various other hosts), B. graminicola sp. nov. (on Poa spp. as principal hosts, but also on various other hosts), and B. hordei sp. nov. (on Hordeum spp.). Synonyms were assessed, some were lectotypified, and questionable names previously associated with powdery mildew on monocots were discussed although their identities remained unresolved. Keys to the described species were developed.
Keywords: Ascomycota; E3 ubiquitinprotein ligase; Poaceae; typification; unnamed orthologous genes.
2021, by The Mycological Society of Japan, © 2021 Her Majesty the Queen in Right of Canada.
Conflict of interest statement
none
Figures







Similar articles
-
Multi-locus phylogeny and taxonomy of an unresolved, heterogeneous species complex within the genus Golovinomyces (Ascomycota, Erysiphales), including G. ambrosiae, G. circumfusus and G. spadiceus.BMC Microbiol. 2020 Mar 5;20(1):51. doi: 10.1186/s12866-020-01731-9. BMC Microbiol. 2020. PMID: 32138640 Free PMC article.
-
Systematics and phylogenetics of Indo-Pacific Luciolinae fireflies (Coleoptera: Lampyridae) and the description of new genera.Zootaxa. 2013;3653:1-162. doi: 10.11646/zootaxa.3653.1.1. Zootaxa. 2013. PMID: 25340191
-
A polyphasic approach leading to the revision of the genus Planktothrix (Cyanobacteria) and its type species, P. agardhii, and proposal for integrating the emended valid botanical taxa, as well as three new species, Planktothrix paucivesiculata sp. nov.ICNP, Planktothrix tepida sp. nov.ICNP, and Planktothrix serta sp. nov.ICNP, as genus and species names with nomenclatural standing under the ICNP.Syst Appl Microbiol. 2015 May;38(3):141-58. doi: 10.1016/j.syapm.2015.02.004. Epub 2015 Feb 24. Syst Appl Microbiol. 2015. PMID: 25757799
-
Monograph on the Cillaeinae (Coleoptera: Nitidulidae) from the Australian Region with comments on the taxonomy of the subfamily.Zootaxa. 2022 Feb 23;5103(1):1-133. doi: 10.11646/zootaxa.5103.1.1. Zootaxa. 2022. PMID: 35391053
-
Combating powdery mildew: Advances in molecular interactions between Blumeria graminis f. sp. tritici and wheat.Front Plant Sci. 2022 Dec 16;13:1102908. doi: 10.3389/fpls.2022.1102908. eCollection 2022. Front Plant Sci. 2022. PMID: 36589137 Free PMC article. Review.
Cited by
-
A powdery mildew core effector protein targets the host endosome tethering complexes HOPS and CORVET in barley.Plant Physiol. 2025 Mar 28;197(4):kiaf067. doi: 10.1093/plphys/kiaf067. Plant Physiol. 2025. PMID: 39973312 Free PMC article.
-
Beyond Nuclear Ribosomal DNA Sequences: Evolution, Taxonomy, and Closest Known Saprobic Relatives of Powdery Mildew Fungi (Erysiphaceae) Inferred From Their First Comprehensive Genome-Scale Phylogenetic Analyses.Front Microbiol. 2022 Jun 9;13:903024. doi: 10.3389/fmicb.2022.903024. eCollection 2022. Front Microbiol. 2022. PMID: 35756050 Free PMC article.
-
Population genomics and molecular epidemiology of wheat powdery mildew in Europe.PLoS Biol. 2025 May 2;23(5):e3003097. doi: 10.1371/journal.pbio.3003097. eCollection 2025 May. PLoS Biol. 2025. PMID: 40315179 Free PMC article.
-
Site-specific analysis reveals candidate cross-kingdom small RNAs, tRNA and rRNA fragments, and signs of fungal RNA phasing in the barley-powdery mildew interaction.Mol Plant Pathol. 2023 Jun;24(6):570-587. doi: 10.1111/mpp.13324. Epub 2023 Mar 14. Mol Plant Pathol. 2023. PMID: 36917011 Free PMC article.
-
Haplotype Analysis Sheds Light on the Genetic Evolution of the Powdery Mildew Resistance Locus Pm60 in Triticum Species.Pathogens. 2023 Feb 2;12(2):241. doi: 10.3390/pathogens12020241. Pathogens. 2023. PMID: 36839513 Free PMC article.
References
-
- Adhikari, M.K. (2017). Researches on the Nepalese mycoflora 3: Erysiphales from Nepal Kathmandu, Nepal: KamalaAdhikari, Kamala S..
-
- Ahmad, N., Agarwal, D., Bambawal, O., & Puzari, K. (2007). Erysiphaceae of India. Monographic Treatment. New Delhi: Venus Printers and Publishers.
-
- Amano, K. (1986). Host Range and Geographical Distribution of the Powdery Mildew Fungi. Japan Scientific Societies Press.
-
- Blumer, S. (1933).Die Erysiphaceen Mitteleuropas mit besonderer Berucksichtigung der Schweiz. Beiträge zur Kryptogamenflora der Schweiz., 7 (1), 483 pp, 167 figs.
-
- Blumer, S. (1967). Echte Mehltaupilze Erysiphaceae: Ein Bestimmungsbuch für die in Europa vorkommenden Arten: Fischer.
LinkOut - more resources
Full Text Sources
Miscellaneous
