Recombinant production, purification, and biochemical characterization of a novel L-lactate dehydrogenase from Bacillus cereus NRC1 and inhibition study of mangiferin
- PMID: 37091329
- PMCID: PMC10117910
- DOI: 10.3389/fbioe.2023.1165465
Recombinant production, purification, and biochemical characterization of a novel L-lactate dehydrogenase from Bacillus cereus NRC1 and inhibition study of mangiferin
Abstract
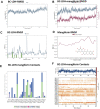
Lactate dehydrogenase (LDH, EC 1.1.1.27) is one of the vital glycolytic conditions, especially during anaerobic conditions. It is a significant diagnostic, prognostic, and monitoring biomarker parameter. A 950-bp DNA fragment containing the gene (LDH) encoding LDH was amplified from Bacillus cereus NRC1. The deduced amino acid sequence reveals that B. cereus LDH (Bc-LDH) is highly homologous to the LDHs of Bacillus organisms. All LDH enzymes have a significant degree of conservation in their active site and several additional domains with unidentified functions. The gene for LDH, which catalyzes lactate synthesis, was cloned, sequenced (accession number: LC706200.1), and expressed in Escherichia coli BL21 (DE3). In this investigation, Bc-LDH was purified to homogeneity with a specific activity of 22.7 units/mg protein and a molecular weight of 35 kDa. It works optimally at pH 8.0. The purified enzyme was inhibited by FeCl2, CuCl2, ZnCl2, and NiCl, whereas CoCl2 was found to boost the activity of Bc-LDH. The molecular docking of the 3D model of the Bc-LDH structure with a natural inhibitor, mangiferin, demonstrated excellent LDH inhibition, with a free binding energy of -10.2 kcal/mol. Moreover, mangiferin is a potent Bc-LDH inhibitor that inhibits Bc-LDH competitively and has one binding site with a Ki value of 0.075 mM. The LDH-mangiferin interaction exhibits a low RMSF value (>1.5 Å), indicating a stable contact at the residues. This study will pave the way for more studies to improve the understanding of mangiferin, which could be considered an intriguing candidate for creating novel and improved LDH inhibitors.
Keywords: Bacillus cereus; characterization; cloning; lactate dehydrogenase; mangiferin; molecular docking.
Copyright © 2023 Esa, El-Sayed, El-Khonezy and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



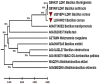

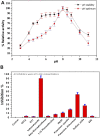



References
-
- Abosereh N. A., Salim R. G., El-Sayed A. F., Hammad M. A., Elsayed G. M. (2022). In-vitro biodegradation of Glyphosate using genetically improved bacterial isolates from highly polluted wastewater. Egypt. J. Chem. 65, 0–681. 10.21608/EJCHEM.2022.141571.6194 - DOI
LinkOut - more resources
Full Text Sources

