Computational and cellular exploration of the protein-protein interaction between Vibrio fischeri STAS domain protein SypA and serine kinase SypE
- PMID: 37091830
- PMCID: PMC10120452
- DOI: 10.1080/19420889.2023.2203626
Computational and cellular exploration of the protein-protein interaction between Vibrio fischeri STAS domain protein SypA and serine kinase SypE
Abstract
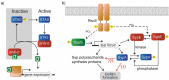
Anti-sigma factor antagonists SpoIIAA and RsbV from Bacillus subtilis are the archetypes for single-domain STAS proteins in bacteria. The structures and mechanisms of these proteins along with their cognate anti-sigma factors have been well studied. SpoIIAA and RsbV utilize a partner-switching mechanism to regulate gene expression through protein-protein interactions to control the activity of their downstream anti-sigma factor partners. The Vibrio fischeri STAS domain protein SypA is also proposed to employ a partner-switching mechanism with its partner SypE, a serine kinase/phosphatase that controls SypA's phosphorylation state. However, this regulation appears opposite to the canonical pathway, with SypA being the more downstream component rather than SypE. Here we explore the commonalities and differences between SypA and the canonical single-domain STAS proteins SpoIIAA and RsbV. We use a combination of AlphaFold 2 structure predictions and computational modeling to investigate the SypA-SypE binding interface. We then test a subset of our predictions in V.fischeri by generating and expressing SypA variants. Our findings suggest that, while SypA shares many sequence and structural traits with anti-sigma factor antagonist STAS domain proteins, there are significant differences that may account for SypA's distinct regulatory output.
Keywords: Anti-sigma factor antagonists; STAS; Vibrio; biofilm; phosphorylation; protein-protein interaction.
© 2023 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures




Similar articles
-
The response regulator SypE controls biofilm formation and colonization through phosphorylation of the syp-encoded regulator SypA in Vibrio fischeri.Mol Microbiol. 2013 Feb;87(3):509-25. doi: 10.1111/mmi.12109. Epub 2012 Dec 11. Mol Microbiol. 2013. PMID: 23171087 Free PMC article.
-
Assessing the function of STAS domain protein SypA in Vibrio fischeri using a comparative analysis.Front Microbiol. 2015 Jul 28;6:760. doi: 10.3389/fmicb.2015.00760. eCollection 2015. Front Microbiol. 2015. PMID: 26284045 Free PMC article.
-
Inhibition of SypG-induced biofilms and host colonization by the negative regulator SypE in Vibrio fischeri.PLoS One. 2013;8(3):e60076. doi: 10.1371/journal.pone.0060076. Epub 2013 Mar 28. PLoS One. 2013. PMID: 23555890 Free PMC article.
-
STAS Domain Only Proteins in Bacterial Gene Regulation.Front Cell Infect Microbiol. 2021 Jun 21;11:679982. doi: 10.3389/fcimb.2021.679982. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34235094 Free PMC article. Review.
-
STAS domain structure and function.Cell Physiol Biochem. 2011;28(3):407-22. doi: 10.1159/000335104. Epub 2011 Nov 16. Cell Physiol Biochem. 2011. PMID: 22116355 Free PMC article. Review.
Cited by
-
Lighting the way: how the Vibrio fischeri model microbe reveals the complexity of Earth's "simplest" life forms.J Bacteriol. 2024 May 23;206(5):e0003524. doi: 10.1128/jb.00035-24. Epub 2024 May 2. J Bacteriol. 2024. PMID: 38695522 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
