Non-structural genes of novel lemur adenoviruses reveal codivergence of virus and host
- PMID: 37091898
- PMCID: PMC10121206
- DOI: 10.1093/ve/vead024
Non-structural genes of novel lemur adenoviruses reveal codivergence of virus and host
Abstract
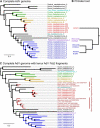
Adenoviruses (AdVs) are important human and animal pathogens and are frequently used as vectors for gene therapy and vaccine delivery. Surprisingly, there are only scant data regarding primate AdV origin and evolution, especially in the most basal primate hosts. We detect and sequence AdVs from faeces of two Madagascan lemur species. Complete genome sequence analyses define a new AdV species with a particularly large gene encoding a protein of unknown function in the early gene region 3. Unexpectedly, the new AdV species is not most similar to human or other simian AdVs but to bat adenovirus C. Genome characterisation shows signals of virus-host codivergence in non-structural genes, which show lower diversity than structural genes. Outside a lemur species mixing zone, recombination less frequently separates structural genes, as in human adenovirus C. The evolutionary history of lemur AdVs likely involves both a host switch and codivergence with the lemur hosts.
Keywords: adenovirus; cospeciation; host-switch; primate; prosimian.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures







References
-
- Ai L. et al. (2022) ‘Genomic Characteristics and Pathogenicity of a New Bat Adenoviruses Strains that Was Isolated in at Sites along the Southeastern Coasts of the P. R. of China from 2015 to 2019’, Virus Research, 308: 198653. - PubMed
-
- Andriamandimbiarisoa L. et al. (2015) ‘Habitat Corridor Utilization by the Gray Mouse Lemur, Microcebus Murinus, in the Littoral Forest Fragments of Southeastern Madagascar’, Madagascar Conservation and Development 10: 144–50.
Grants and funding
LinkOut - more resources
Full Text Sources

