Multi-omic analyses in immune cell development with lessons learned from T cell development
- PMID: 37091971
- PMCID: PMC10118026
- DOI: 10.3389/fcell.2023.1163529
Multi-omic analyses in immune cell development with lessons learned from T cell development
Abstract
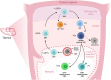
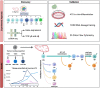
Traditionally, flow cytometry has been the preferred method to characterize immune cells at the single-cell level. Flow cytometry is used in immunology mostly to measure the expression of identifying markers on the cell surface, but-with good antibodies-can also be used to assess the expression of intracellular proteins. The advent of single-cell RNA-sequencing has paved the road to study immune development at an unprecedented resolution. Single-cell RNA-sequencing studies have not only allowed us to efficiently chart the make-up of heterogeneous tissues, including their most rare cell populations, it also increasingly contributes to our understanding how different omics modalities interplay at a single cell resolution. Particularly for investigating the immune system, this means that these single-cell techniques can be integrated to combine and correlate RNA and protein data at the single-cell level. While RNA data usually reveals a large heterogeneity of a given population identified solely by a combination of surface protein markers, the integration of different omics modalities at a single cell resolution is expected to greatly contribute to our understanding of the immune system.
Keywords: T cells; multi‐omics; scRNA‐seq; spectral flow cytometry; thymus.
Copyright © 2023 Cordes, Pike-Overzet, Van Den Akker, Staal and Canté-Barrett.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
DEMOC: a deep embedded multi-omics learning approach for clustering single-cell CITE-seq data.Brief Bioinform. 2022 Sep 20;23(5):bbac347. doi: 10.1093/bib/bbac347. Brief Bioinform. 2022. PMID: 36047285
-
Integration of scATAC-Seq with scRNA-Seq Data.Methods Mol Biol. 2023;2584:293-310. doi: 10.1007/978-1-0716-2756-3_15. Methods Mol Biol. 2023. PMID: 36495457
-
Single-cell multi-omic topic embedding reveals cell-type-specific and COVID-19 severity-related immune signatures.bioRxiv [Preprint]. 2023 Jun 1:2023.01.31.526312. doi: 10.1101/2023.01.31.526312. bioRxiv. 2023. Update in: Cell Rep Methods. 2023 Aug 18;3(8):100563. doi: 10.1016/j.crmeth.2023.100563. PMID: 36778483 Free PMC article. Updated. Preprint.
-
Single-cell RNA sequencing in breast cancer: Understanding tumor heterogeneity and paving roads to individualized therapy.Cancer Commun (Lond). 2020 Aug;40(8):329-344. doi: 10.1002/cac2.12078. Epub 2020 Jul 12. Cancer Commun (Lond). 2020. PMID: 32654419 Free PMC article. Review.
-
Intricacies of single-cell multi-omics data integration.Trends Genet. 2022 Feb;38(2):128-139. doi: 10.1016/j.tig.2021.08.012. Epub 2021 Sep 21. Trends Genet. 2022. PMID: 34561102 Review.
Cited by
-
Using combined single-cell gene expression, TCR sequencing and cell surface protein barcoding to characterize and track CD4+ T cell clones from murine tissues.Front Immunol. 2023 Oct 12;14:1241283. doi: 10.3389/fimmu.2023.1241283. eCollection 2023. Front Immunol. 2023. PMID: 37901204 Free PMC article.
-
Systematic review and quantitative meta-analysis of age-dependent human T-lymphocyte homeostasis.Front Immunol. 2025 Jan 27;16:1475871. doi: 10.3389/fimmu.2025.1475871. eCollection 2025. Front Immunol. 2025. PMID: 39931065 Free PMC article.
-
The recombinase activating genes: architects of immune diversity during lymphocyte development.Front Immunol. 2023 Jul 11;14:1210818. doi: 10.3389/fimmu.2023.1210818. eCollection 2023. Front Immunol. 2023. PMID: 37497222 Free PMC article. Review.
-
Deciphering the Complexities of Adult Human Steady State and Stress-Induced Hematopoiesis: Progress and Challenges.Int J Mol Sci. 2025 Jan 14;26(2):671. doi: 10.3390/ijms26020671. Int J Mol Sci. 2025. PMID: 39859383 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources

