Inheritance of social dominance is associated with global sperm DNA methylation in inbred male mice
- PMID: 37092005
- PMCID: PMC10120999
- DOI: 10.1093/cz/zoac030
Inheritance of social dominance is associated with global sperm DNA methylation in inbred male mice
Abstract
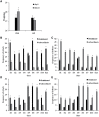
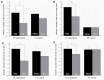
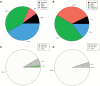
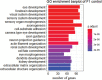
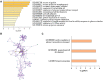
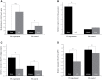
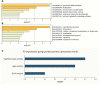
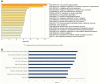
Dominance relationships between males and their associated traits are usually heritable and have implications for sexual selection in animals. In particular, social dominance and its related male pheromones are heritable in inbred mice; thus, we wondered whether epigenetic changes due to altered levels of DNA methylation determine inheritance. Here, we used C57BL/6 male mice to establish a social dominance-subordination relationship through chronic dyadic encounters, and this relationship and pheromone covariation occurred in their offspring, indicative of heritability. Through transcriptome sequencing and whole-genome DNA methylation profiling of the sperm of both generations, we found that differential methylation of many genes was induced by social dominance-subordination in sires and could be passed on to the offspring. These methylated genes were mainly related to growth and development processes, neurodevelopment, and cellular transportation. The expression of the genes with similar functions in whole-genome methylation/bisulfite sequencing was also differentiated by social dominance-subordination, as revealed by RNA-seq. In particular, the gene Dennd1a, which regulates neural signaling, was differentially methylated and expressed in the sperm and medial prefrontal cortex in paired males before and after dominance-subordination establishment, suggesting the potential epigenetic control and inheritance of social dominance-related aggression. We suggest that social dominance might be passed on to male offspring through sperm DNA methylation and that the differences could potentially affect male competition in offspring by affecting the development of the nervous system.
Keywords: WGBS; aggression; epigenetic inheritance; pheromone; social hierarchy.
© The Author(s) 2022. Published by Oxford University Press on behalf of Editorial Office, Current Zoology.
Conflict of interest statement
The Authors declare that there is no conflict of interest.
Figures


Similar articles
-
Transgenerational diabetogenic effects of preconception exposure to inorganic arsenic in C57BL/6 mice are associated with dysregulation of DNA methylation and gene expression in G1 and G2 offspring.Arch Toxicol. 2025 Jun 12. doi: 10.1007/s00204-025-04107-y. Online ahead of print. Arch Toxicol. 2025. PMID: 40504410
-
Individuality and Transgenerational Inheritance of Social Dominance and Sex Pheromones in Isogenic Male Mice.J Exp Zool B Mol Dev Evol. 2016 Jun;326(4):225-36. doi: 10.1002/jez.b.22681. Epub 2016 Jun 10. J Exp Zool B Mol Dev Evol. 2016. PMID: 27283352
-
Increased paternal corticosterone exposure influences offspring behaviour and expression of urinary pheromones.BMC Biol. 2023 Sep 5;21(1):186. doi: 10.1186/s12915-023-01678-z. BMC Biol. 2023. PMID: 37667240 Free PMC article.
-
Age-associated epigenetic changes in mammalian sperm: implications for offspring health and development.Hum Reprod Update. 2023 Jan 5;29(1):24-44. doi: 10.1093/humupd/dmac033. Hum Reprod Update. 2023. PMID: 36066418 Free PMC article. Review.
-
Environmental epigenetic inheritance through gametes and implications for human reproduction.Hum Reprod Update. 2015 Mar-Apr;21(2):194-208. doi: 10.1093/humupd/dmu061. Epub 2014 Nov 21. Hum Reprod Update. 2015. PMID: 25416302 Review.
Cited by
-
Dopamine D2 receptors in the dorsomedial prefrontal cortex modulate social hierarchy in male mice.Curr Zool. 2022 Nov 7;69(6):682-693. doi: 10.1093/cz/zoac087. eCollection 2023 Dec. Curr Zool. 2022. PMID: 37876636 Free PMC article.
References
-
- Andersson M, Simmons LW, 2006. Sexual selection and mate choice. Trend Ecol Evol 21:296–302. - PubMed
LinkOut - more resources
Full Text Sources
