MKRN3 inhibits puberty onset via interaction with IGF2BP1 and regulation of hypothalamic plasticity
- PMID: 37092553
- PMCID: PMC10243807
- DOI: 10.1172/jci.insight.164178
MKRN3 inhibits puberty onset via interaction with IGF2BP1 and regulation of hypothalamic plasticity
Abstract
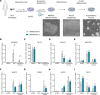
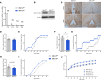
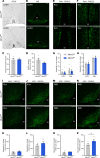
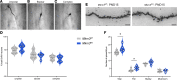
Makorin ring finger protein 3 (MKRN3) was identified as an inhibitor of puberty initiation with the report of loss-of-function mutations in association with central precocious puberty. Consistent with this inhibitory role, a prepubertal decrease in Mkrn3 expression was observed in the mouse hypothalamus. Here, we investigated the mechanisms of action of MKRN3 in the central regulation of puberty onset. We showed that MKRN3 deletion in hypothalamic neurons derived from human induced pluripotent stem cells was associated with significant changes in expression of genes controlling hypothalamic development and plasticity. Mkrn3 deletion in a mouse model led to early puberty onset in female mice. We found that Mkrn3 deletion increased the number of dendritic spines in the arcuate nucleus but did not alter the morphology of GnRH neurons during postnatal development. In addition, we identified neurokinin B (NKB) as an Mkrn3 target. Using proteomics, we identified insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) as another target of MKRN3. Interactome analysis revealed that IGF2BP1 interacted with MKRN3, along with several members of the polyadenylate-binding protein family. Our data show that one of the mechanisms by which MKRN3 inhibits pubertal initiation is through regulation of prepubertal hypothalamic development and plasticity, as well as through effects on NKB and IGF2BP1.
Keywords: Endocrinology; Mouse models; Neurodevelopment; Neuroendocrine regulation; Neuroscience.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

