Conformational transition of the Ixodes ricinus salivary serpin Iripin-4
- PMID: 37092969
- PMCID: PMC10167670
- DOI: 10.1107/S2059798323002322
Conformational transition of the Ixodes ricinus salivary serpin Iripin-4
Abstract
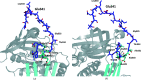
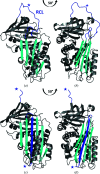
Iripin-4, one of the many salivary serpins from Ixodes ricinus ticks with an as-yet unexplained function, crystallized in two different structural conformations, namely the native partially relaxed state and the cleaved serpin. The native structure was solved at a resolution of 2.3 Å and the structure of the cleaved conformation was solved at 2.0 Å resolution. Furthermore, structural changes were observed when the reactive-centre loop transitioned from the native conformation to the cleaved conformation. In addition to this finding, it was confirmed that Glu341 represents a primary substrate-recognition site for the inhibitory mechanism. The presence of glutamate instead of the typical arginine in the P1 recognition site of all structurally characterized I. ricinus serpins (PDB entries 7b2t, 7pmu and 7ahp), except for the tyrosine in the P1 site of Iripin-2 (formerly IRS-2; PDB entry 3nda), would explain the absence of inhibition of the tested proteases that cleave their substrate after arginine. Further research on Iripin-4 should focus on functional analysis of this interesting serpin.
Keywords: Iripin-4; Ixodes ricinus; X-ray structure; cleaved conformation; native conformation; serpins.
open access.
Figures



Similar articles
-
Structural and biochemical characterization of the novel serpin Iripin-5 from Ixodes ricinus.Acta Crystallogr D Struct Biol. 2021 Sep 1;77(Pt 9):1183-1196. doi: 10.1107/S2059798321007920. Epub 2021 Aug 23. Acta Crystallogr D Struct Biol. 2021. PMID: 34473088 Free PMC article.
-
Ixodes ricinus Salivary Serpin Iripin-8 Inhibits the Intrinsic Pathway of Coagulation and Complement.Int J Mol Sci. 2021 Aug 31;22(17):9480. doi: 10.3390/ijms22179480. Int J Mol Sci. 2021. PMID: 34502392 Free PMC article.
-
Iripin-1, a new anti-inflammatory tick serpin, inhibits leukocyte recruitment in vivo while altering the levels of chemokines and adhesion molecules.Front Immunol. 2023 Jan 23;14:1116324. doi: 10.3389/fimmu.2023.1116324. eCollection 2023. Front Immunol. 2023. PMID: 36756125 Free PMC article.
-
A protein family under 'stress' - serpin stability, folding and misfolding.Front Biosci. 2005 Jan 1;10:288-99. doi: 10.2741/1528. Print 2005 Jan 1. Front Biosci. 2005. PMID: 15574369 Review.
-
Serpins flex their muscle: II. Structural insights into target peptidase recognition, polymerization, and transport functions.J Biol Chem. 2010 Aug 6;285(32):24307-12. doi: 10.1074/jbc.R110.141408. Epub 2010 May 24. J Biol Chem. 2010. PMID: 20498368 Free PMC article. Review.
Cited by
-
Genome sequences of four Ixodes species expands understanding of tick evolution.BMC Biol. 2025 Jan 21;23(1):17. doi: 10.1186/s12915-025-02121-1. BMC Biol. 2025. PMID: 39838418 Free PMC article.
References
-
- Boulanger, N., Boyer, P., Talagrand-Reboul, E. & Hansmann, Y. (2019). Med. Mal. Infect. 49, 87–97. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

