Initiator AUGs Are Discriminated from Elongator AUGs Predominantly through mRNA Accessibility in C. crescentus
- PMID: 37092987
- PMCID: PMC10210977
- DOI: 10.1128/jb.00420-22
Initiator AUGs Are Discriminated from Elongator AUGs Predominantly through mRNA Accessibility in C. crescentus
Abstract
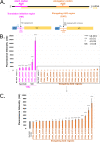
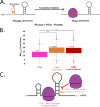
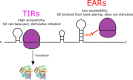
The initiation of translation in bacteria is thought to occur upon base pairing between the Shine-Dalgarno (SD) site in the mRNA and the anti-SD site in the rRNA. However, in many bacterial species, such as Caulobacter crescentus, a minority of mRNAs have SD sites. To examine the functional importance of SD sites in C. crescentus, we analyzed the transcriptome and found that more SD sites exist in the coding sequence than in the preceding start codons. To examine the function of SD sites in initiation, we designed a series of mutants with altered ribosome accessibility and SD content in translation initiation regions (TIRs) and in elongator AUG regions (EARs). A lack of mRNA structure content is required for initiation in TIRs, and, when introduced into EARs, can stimulate initiation, thereby suggesting that low mRNA structure content is a major feature that is required for initiation. SD sites appear to stimulate initiation in TIRs, which generally lack structure content, but SD sites only stimulate initiation in EARs if RNA secondary structures are destabilized. Taken together, these results suggest that the difference in secondary structure between TIRs and EARs directs ribosomes to start codons where SD base pairing can tune the efficiency of initiation, but SDs in EARs do not stimulate initiation, as they are blocked by stable secondary structures. This highlights the importance of studying translation initiation mechanisms in diverse bacterial species. IMPORTANCE Start codon selection is an essential process that is thought to occur via the base pairing of the rRNA to the SD site in the mRNA. This model is based on studies in E. coli, yet whole-genome sequencing revealed that SD sites are absent at start codons in many species. By examining the transcriptome of C. crescentus, we found more SD-AUG pairs in the CDS of mRNAs than preceding start codons, yet these internal sites do not initiate. Instead, start codon regions have lower mRNA secondary structure content than do internal SD-AUG regions. Therefore, we find that start codon selection is not controlled by the presence of SD sites, which tune initiation efficiency, but by lower mRNA structure content surrounding the start codon.
Keywords: Caulobacter crescentus; mRNA structure; ribosomes; translation initiation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

