The Periplasmic Tail-Specific Protease, Tsp, Is Essential for Secondary Differentiation in Chlamydia trachomatis
- PMID: 37092988
- PMCID: PMC10210983
- DOI: 10.1128/jb.00099-23
The Periplasmic Tail-Specific Protease, Tsp, Is Essential for Secondary Differentiation in Chlamydia trachomatis
Abstract
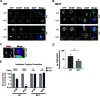
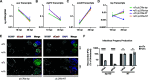
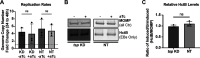
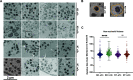
The obligate intracellular human pathogen Chlamydia trachomatis (Ctr) undergoes a complex developmental cycle in which the bacterium differentiates between two functionally and morphologically distinct forms: the elementary body (EB) and the reticulate body (RB). The EB is the smaller, infectious, nondividing form which initiates infection of a susceptible host cell, whereas the RB is the larger, non-infectious form which replicates within a membrane-bound vesicle called an inclusion. The mechanism(s) which drives differentiation between these developmental forms is poorly understood. Bulk protein turnover is likely required for chlamydial differentiation given the significant differences in the protein repertoires and functions of the EB and RB. We hypothesize that periplasmic protein turnover is also critical for the reorganization of an RB into an EB, referred to as secondary differentiation. Ct441 is a periplasmic protease ortholog of tail-specific proteases (i.e., Tsp, Prc) and is expressed in Ctr during secondary differentiation. We investigated the effect of altering Tsp expression on developmental cycle progression. Through assessment of bacterial morphology and infectious progeny production, we found that both overexpression and CRISPR interference/dCas9 (CRISPRi)-mediated knockdown of Tsp negatively impacted chlamydial development through different mechanisms. We also confirmed that catalytic activity is required for the negative effect of overexpression and confirmed the effect of the mutation in in vitro assays. Electron microscopic assessments during knockdown experiments revealed a defect in EB morphology, directly linking Tsp function to secondary differentiation. These data implicate Ct441/Tsp as a critical factor in secondary differentiation. IMPORTANCE The human pathogen Chlamydia trachomatis is the leading cause of preventable infectious blindness and bacterial sexually transmitted infections worldwide. This pathogen has a unique developmental cycle that alternates between distinct forms. However, the key processes of chlamydial development remain obscure. Uncovering the mechanisms of differentiation between its metabolically and functionally distinct developmental forms may foster the discovery of novel Chlamydia-specific therapeutics and limit development of resistant bacterial populations derived from the clinical use of broad-spectrum antibiotics. In this study, we investigate chlamydial tail-specific protease (Tsp) and its function in chlamydial growth and development. Our work implicates Tsp as essential to chlamydial developmental cycle progression and indicates that Tsp is a potential drug target for Chlamydia infections.
Keywords: Chlamydia; Prc; Tsp; development; differentiation; periplasm; protease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Type III Secretion in Chlamydia.Microbiol Mol Biol Rev. 2023 Sep 26;87(3):e0003423. doi: 10.1128/mmbr.00034-23. Epub 2023 Jun 26. Microbiol Mol Biol Rev. 2023. PMID: 37358451 Free PMC article. Review.
-
The ClpX and ClpP2 Orthologs of Chlamydia trachomatis Perform Discrete and Essential Functions in Organism Growth and Development.mBio. 2020 Sep 1;11(5):e02016-20. doi: 10.1128/mBio.02016-20. mBio. 2020. PMID: 32873765 Free PMC article.
-
Tag-Dependent Substrate Selection of ClpX Underlies Secondary Differentiation of Chlamydia trachomatis.mBio. 2022 Oct 26;13(5):e0185822. doi: 10.1128/mbio.01858-22. Epub 2022 Sep 26. mBio. 2022. PMID: 36154190 Free PMC article.
-
The T3SS structural and effector genes of Chlamydia trachomatis are expressed in distinct phenotypic cell forms.Front Cell Infect Microbiol. 2025 May 8;15:1579247. doi: 10.3389/fcimb.2025.1579247. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40406518 Free PMC article.
-
One Face of Chlamydia trachomatis: The Infectious Elementary Body.Curr Top Microbiol Immunol. 2018;412:35-58. doi: 10.1007/82_2016_12. Curr Top Microbiol Immunol. 2018. PMID: 27197644 Review.
Cited by
-
Type III Secretion in Chlamydia.Microbiol Mol Biol Rev. 2023 Sep 26;87(3):e0003423. doi: 10.1128/mmbr.00034-23. Epub 2023 Jun 26. Microbiol Mol Biol Rev. 2023. PMID: 37358451 Free PMC article. Review.
-
Overexpressing the ClpC AAA+ unfoldase accelerates developmental cycle progression in Chlamydia trachomatis.mBio. 2025 Jan 8;16(1):e0287024. doi: 10.1128/mbio.02870-24. Epub 2024 Nov 22. mBio. 2025. PMID: 39576108 Free PMC article.
-
Molecular pathogenesis of Chlamydia trachomatis.Front Cell Infect Microbiol. 2023 Oct 18;13:1281823. doi: 10.3389/fcimb.2023.1281823. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37920447 Free PMC article. Review.
-
Identification of the alternative sigma factor regulons of Chlamydia trachomatis using multiplexed CRISPR interference.mSphere. 2023 Oct 24;8(5):e0039123. doi: 10.1128/msphere.00391-23. Epub 2023 Sep 25. mSphere. 2023. PMID: 37747235 Free PMC article.
-
Late gene regulation by the alternative sigma factors of Chlamydia trachomatis.mSystems. 2025 Jul 22;10(7):e0029225. doi: 10.1128/msystems.00292-25. Epub 2025 Jun 12. mSystems. 2025. PMID: 40503901 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

