Specific Residues in the C-Terminal Domain of the Human Metapneumovirus Phosphoprotein Are Indispensable for Formation of Viral Replication Centers and Regulation of the Function of the Viral Polymerase Complex
- PMID: 37092993
- PMCID: PMC10231248
- DOI: 10.1128/jvi.00030-23
Specific Residues in the C-Terminal Domain of the Human Metapneumovirus Phosphoprotein Are Indispensable for Formation of Viral Replication Centers and Regulation of the Function of the Viral Polymerase Complex
Abstract
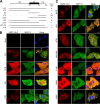
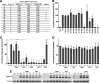
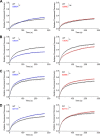
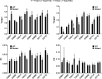
Human metapneumovirus (HMPV) is a negative-strand RNA virus that frequently causes respiratory tract infections in infants, the elderly, and the immunocompromised. A hallmark of HMPV infection is the formation of membraneless, liquid-like replication and transcription centers in the cytosol termed inclusion bodies (IBs). The HMPV phosphoprotein (P) and nucleoprotein (N) are the minimal viral proteins necessary to form IB-like structures, and both proteins are required for the viral polymerase to synthesize RNA during infection. HMPV P is a homotetramer with regions of intrinsic disorder and has several known and predicted phosphorylation sites of unknown function. In this study, we found that the P C-terminal intrinsically disordered domain (CTD) must be present to facilitate IB formation with HMPV N, while either the N-terminal intrinsically disordered domain or the central oligomerization domain was dispensable. Alanine substitution at a single tyrosine residue within the CTD abrogated IB formation and reduced coimmunoprecipitation with HMPV N. Mutations to C-terminal phosphorylation sites revealed a potential role for phosphorylation in regulating RNA synthesis and P binding partners within IBs. Phosphorylation mutations which reduced RNA synthesis in a reporter assay produced comparable results in a recombinant viral rescue system, measured as an inability to produce infectious viral particles with genomes containing these single P mutations. This work highlights the critical role HMPV P plays in facilitating a key step of the viral life cycle and reveals the potential role for phosphorylation in regulating the function of this significant viral protein. IMPORTANCE Human metapneumovirus (HMPV) infects global populations, with severe respiratory tract infections occurring in infants, the elderly, and the immunocompromised. There are currently no FDA-approved therapeutics available to prevent or treat HMPV infection. Therefore, understanding how HMPV replicates is vital for the identification of novel targets for therapeutic development. During HMPV infection, viral RNA synthesis proteins localize to membraneless structures called inclusion bodies (IBs), which are sites of genome replication and transcription. The HMPV phosphoprotein (P) is necessary for IBs to form and for the virus to synthesize RNA, but it is not known how this protein contributes to IB formation or if it is capable of regulating viral replication. We show that the C-terminal domain of P is the location of a molecular interaction driving IB formation and contains potential phosphorylation sites where amino acid charge regulates the function of the viral polymerase complex.
Keywords: inclusion bodies; phosphoprotein; pneumovirus; protein phosphorylation; replication complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Afonso CL, Amarasinghe GK, Bányai K, Bào Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand FX, Briese T, Bukreyev A, Calisher CH, Chandran K, Chéng J, Clawson AN, Collins PL, Dietzgen RG, Dolnik O, Domier LL, Dürrwald R, Dye JM, Easton AJ, Ebihara H, Farkas SL, Freitas-Astúa J, Formenty P, Fouchier RA, Fù Y, Ghedin E, Goodin MM, Hewson R, Horie M, Hyndman TH, Jiāng D, Kitajima EW, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lenardon S, Leroy EM, Li CX, Lin XD, Liú L, Longdon B, Marton S, Maisner A, Mühlberger E, Netesov SV, Nowotny N, et al. 2016. Taxonomy of the order Mononegavirales: update 2016. Arch Virol 161:2351–2360. doi: 10.1007/s00705-016-2880-1. - DOI - PMC - PubMed
-
- Wang X, Li Y, Deloria-Knoll M, Madhi SA, Cohen C, Ali A, Basnet S, Bassat Q, Brooks WA, Chittaganpitch M, Echavarria M, Fasce RA, Goswami D, Hirve S, Homaira N, Howie SRC, Kotloff KL, Khuri-Bulos N, Krishnan A, Lucero MG, Lupisan S, Mira-Iglesias A, Moore DP, Moraleda C, Nunes M, Oshitani H, Owor BE, Polack FP, O’Brien KL, Rasmussen ZA, Rath BA, Salimi V, Scott JAG, Simões EAF, Strand TA, Thea DM, Treurnicht FK, Vaccari LC, Yoshida L-M, Zar HJ, Campbell H, Nair H, Respiratory Virus Global Epidemiology Network . 2021. Global burden of acute lower respiratory infection associated with human metapneumovirus in children under 5 years in 2018: a systematic review and modelling study. Lancet Global Health 9:e33–e43. doi: 10.1016/S2214-109X(21)00218-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

