A Bioinformatics Assessment Indicating Better Outcomes With Breast Cancer Resident, Immunoglobulin CDR3-MMP2 Binding
- PMID: 37093687
- PMCID: PMC10148069
- DOI: 10.21873/cgp.20378
A Bioinformatics Assessment Indicating Better Outcomes With Breast Cancer Resident, Immunoglobulin CDR3-MMP2 Binding
Abstract
Background/aim: The recombination of V, D, and J immunoglobulin (IG) gene segments leads to many variations in the amino acids (AAs) encoded at that site, the complementarity determining region-3 (CDR3). Thus, cancer patients may have varying degrees of CDR3 AA binding specificity for cancer proteases, for example, matrix metalloproteinase 2 (MMP2). MMP2 in breast cancer has been found to contribute to metastasis and is used as a marker for tumor staging. Thus, this report evaluated the tumor resident, patient specific IG CDR3 binding affinities to cancer proteases to test the hypothesis that greater binding affinities would be associated with a better outcome.
Materials and methods: Using two independent bioinformatics tools, we evaluated the IG CDR3-MMP2 binding affinities throughout the cancer genome atlas breast cancer (TCGA-BRCA) dataset.
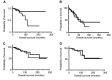
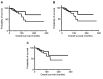
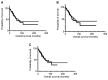
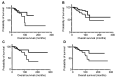
Results: Results indicated that the better the CDR3-MMP2 binding, the better the survival probability. An analogous evaluation for four other proteases, including calpain-1 and thermolysin, displayed no such associations with survival probabilities.
Conclusion: This study is consistent with the possibility that patient IG-cancer protease interactions could impact outcomes and raises the question of whether therapeutic antibody targeting of MMP2 would reduce breast cancer mediated tissue destruction and breast cancer mortality rates.
Keywords: Chemical complementarity bioinformatics; MMP2; breast cancer; immunoglobulin CDR3s.
Copyright © 2023, International Institute of Anticancer Research (Dr. George J. Delinasios), All rights reserved.
Conflict of interest statement
The Authors declare that they have no conflicts of interest.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
