Human CCR6+ Th Cells Show Both an Extended Stable Gradient of Th17 Activity and Imprinted Plasticity
- PMID: 37093875
- PMCID: PMC10463241
- DOI: 10.4049/jimmunol.2200874
Human CCR6+ Th Cells Show Both an Extended Stable Gradient of Th17 Activity and Imprinted Plasticity
Abstract
Th17 cells have been investigated in mice primarily for their contributions to autoimmune diseases. However, the pathways of differentiation of Th17 and related Th cells (type 17 cells) and the structure of the type 17 memory population in humans are not well understood; such understanding is critical for manipulating these cells in vivo. By exploiting differences in levels of surface CCR6, we found that human type 17 memory cells, including individual T cell clonotypes, form an elongated continuum of type 17 character along which cells can be driven by increasing RORγt. This continuum includes cells preserved within the memory pool with potentials that reflect the early preferential activation of multiple over single lineages. The phenotypes and epigenomes of CCR6+ cells are stable across cell divisions under noninflammatory conditions. Nonetheless, activation in polarizing and nonpolarizing conditions can yield additional functionalities, revealing, respectively, both environmentally induced and imprinted mechanisms that contribute differentially across the type 17 continuum to yield the unusual plasticity ascribed to type 17 cells.
Conflict of interest statement
Disclosures
The authors have no financial conflict of interest.
Figures
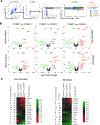

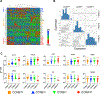
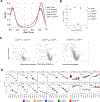




Update of
-
Human CCR6 + Th cells show both an extended stable gradient of Th17 activity and imprinted plasticity.bioRxiv [Preprint]. 2023 Jan 6:2023.01.05.522630. doi: 10.1101/2023.01.05.522630. bioRxiv. 2023. Update in: J Immunol. 2023 Jun 1;210(11):1700-1716. doi: 10.4049/jimmunol.2200874. PMID: 36789418 Free PMC article. Updated. Preprint.
Similar articles
-
Human CCR6 + Th cells show both an extended stable gradient of Th17 activity and imprinted plasticity.bioRxiv [Preprint]. 2023 Jan 6:2023.01.05.522630. doi: 10.1101/2023.01.05.522630. bioRxiv. 2023. Update in: J Immunol. 2023 Jun 1;210(11):1700-1716. doi: 10.4049/jimmunol.2200874. PMID: 36789418 Free PMC article. Updated. Preprint.
-
PLZF regulates CCR6 and is critical for the acquisition and maintenance of the Th17 phenotype in human cells.J Immunol. 2015 May 1;194(9):4350-61. doi: 10.4049/jimmunol.1401093. Epub 2015 Apr 1. J Immunol. 2015. PMID: 25833398 Free PMC article.
-
Increased plasticity of non-classic Th1 cells toward the Th17 phenotype.Mod Rheumatol. 2020 Sep;30(5):930-936. doi: 10.1080/14397595.2019.1667473. Epub 2019 Oct 17. Mod Rheumatol. 2020. PMID: 31512538
-
The role and modulation of CCR6+ Th17 cell populations in rheumatoid arthritis.Cytokine. 2015 Jul;74(1):43-53. doi: 10.1016/j.cyto.2015.02.002. Epub 2015 Mar 28. Cytokine. 2015. PMID: 25828206 Review.
-
CCR6-CCL20 axis as a therapeutic target for autoimmune diseases.Autoimmun Rev. 2021 Jul;20(7):102846. doi: 10.1016/j.autrev.2021.102846. Epub 2021 May 7. Autoimmun Rev. 2021. PMID: 33971346 Review.
Cited by
-
Chemokine positioning determines mutually exclusive roles for their receptors in extravasation of pathogenic human T cells.bioRxiv [Preprint]. 2023 Feb 13:2023.01.25.525561. doi: 10.1101/2023.01.25.525561. bioRxiv. 2023. PMID: 36789428 Free PMC article. Preprint.
-
T cell signatures associated with reduced Chlamydia trachomatis reinfection in a highly exposed cohort.JCI Insight. 2025 Feb 27;10(7):e189388. doi: 10.1172/jci.insight.189388. JCI Insight. 2025. PMID: 40014387 Free PMC article.
-
Altered memory CCR6+ Th17-polarised T-cell function and biology in people with HIV under successful antiretroviral therapy and HIV elite controllers.EBioMedicine. 2024 Sep;107:105274. doi: 10.1016/j.ebiom.2024.105274. Epub 2024 Aug 22. EBioMedicine. 2024. PMID: 39178742 Free PMC article.
-
Staphylococcus aureus-specific TIGIT+ Treg are present in the blood of healthy subjects - a hurdle for vaccination?Front Immunol. 2025 Feb 4;15:1500696. doi: 10.3389/fimmu.2024.1500696. eCollection 2024. Front Immunol. 2025. PMID: 39981298 Free PMC article.
-
Migration arrest and transendothelial trafficking of human pathogenic-like Th17 cells are mediated by differentially positioned chemokines.Nat Commun. 2025 Feb 26;16(1):1978. doi: 10.1038/s41467-025-57002-6. Nat Commun. 2025. PMID: 40000641 Free PMC article.
References
-
- Farber DL, Netea MG, Radbruch A, Rajewsky K, and Zinkernagel RM. 2016. Immunological memory: lessons from the past and a look to the future. Nat Rev Immunol 16: 124–128. - PubMed
-
- Hammarlund E, Lewis MW, Hansen SG, Strelow LI, Nelson JA, Sexton GJ, Hanifin JM, and Slifka MK. 2003. Duration of antiviral immunity after smallpox vaccination. Nat Med 9: 1131–1137. - PubMed
-
- Naniche D, Garenne M, Rae C, Manchester M, Buchta R, Brodine SK, and Oldstone MB. 2004. Decrease in measles virus-specific CD4 T cell memory in vaccinated subjects. J Infect Dis 190: 1387–1395. - PubMed
-
- Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, Shen F, Eaton SM, Gaffen SL, Swain SL, Locksley RM, Haynes L, Randall TD, and Cooper AM. 2007. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol 8: 369–377. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

