Cheating leads to the evolution of multipartite viruses
- PMID: 37093882
- PMCID: PMC10159356
- DOI: 10.1371/journal.pbio.3002092
Cheating leads to the evolution of multipartite viruses
Abstract
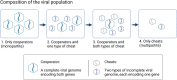
In multipartite viruses, the genome is split into multiple segments, each of which is transmitted via a separate capsid. The existence of multipartite viruses poses a problem, because replication is only possible when all segments are present within the same host. Given this clear cost, why is multipartitism so common in viruses? Most previous hypotheses try to explain how multipartitism could provide an advantage. In so doing, they require scenarios that are unrealistic and that cannot explain viruses with more than 2 multipartite segments. We show theoretically that selection for cheats, which avoid producing a shared gene product, but still benefit from gene products produced by other genomes, can drive the evolution of both multipartite and segmented viruses. We find that multipartitism can evolve via cheating under realistic conditions and does not require unreasonably high coinfection rates or any group-level benefit. Furthermore, the cheating hypothesis is consistent with empirical patterns of cheating and multipartitism across viruses. More broadly, our results show how evolutionary conflict can drive new patterns of genome organisation in viruses and elsewhere.
Copyright: © 2023 Leeks et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist
Figures

Similar articles
-
Theoretical approaches to disclosing the emergence and adaptive advantages of multipartite viruses.Curr Opin Virol. 2018 Dec;33:89-95. doi: 10.1016/j.coviro.2018.07.018. Epub 2018 Aug 16. Curr Opin Virol. 2018. PMID: 30121469 Review.
-
The Curious Strategy of Multipartite Viruses.Annu Rev Virol. 2020 Sep 29;7(1):203-218. doi: 10.1146/annurev-virology-010220-063346. Annu Rev Virol. 2020. PMID: 32991271 Review.
-
Multipartite viruses: adaptive trick or evolutionary treat?NPJ Syst Biol Appl. 2017 Nov 9;3:34. doi: 10.1038/s41540-017-0035-y. eCollection 2017. NPJ Syst Biol Appl. 2017. PMID: 29263796 Free PMC article. Review.
-
Evolutionary dynamics of genome segmentation in multipartite viruses.Proc Biol Sci. 2012 Sep 22;279(1743):3812-9. doi: 10.1098/rspb.2012.1086. Epub 2012 Jul 4. Proc Biol Sci. 2012. PMID: 22764164 Free PMC article.
-
Endemicity and prevalence of multipartite viruses under heterogeneous between-host transmission.PLoS Comput Biol. 2019 Mar 18;15(3):e1006876. doi: 10.1371/journal.pcbi.1006876. eCollection 2019 Mar. PLoS Comput Biol. 2019. PMID: 30883545 Free PMC article.
Cited by
-
Conserved untranslated regions of multipartite viruses: Natural markers of novel viral genomic components and tags of viral evolution.Virus Evol. 2024 Jan 12;10(1):veae004. doi: 10.1093/ve/veae004. eCollection 2024. Virus Evol. 2024. PMID: 38361819 Free PMC article.
-
Relative frequency dynamics and loading of beet necrotic yellow vein virus genomic RNAs during the acquisition by its vector Polymyxa betae.J Virol. 2025 Jan 31;99(1):e0141024. doi: 10.1128/jvi.01410-24. Epub 2024 Dec 16. J Virol. 2025. PMID: 39679720 Free PMC article.
-
New clades of viruses infecting the obligatory biotroph Bremia lactucae representing distinct evolutionary trajectory for viruses infecting oomycetes.Virus Evol. 2024 Jan 5;10(1):veae003. doi: 10.1093/ve/veae003. eCollection 2024. Virus Evol. 2024. PMID: 38361818 Free PMC article.
-
cmv1-Mediated Resistance to CMV in Melon Can Be Overcome by Mixed Infections with Potyviruses.Viruses. 2023 Aug 23;15(9):1792. doi: 10.3390/v15091792. Viruses. 2023. PMID: 37766198 Free PMC article.
-
A phylogenetic approach to comparative genomics.Nat Rev Genet. 2025 Jun;26(6):395-405. doi: 10.1038/s41576-024-00803-0. Epub 2025 Jan 8. Nat Rev Genet. 2025. PMID: 39779997 Free PMC article. Review.
References
-
- Asadulghani M, Ogura Y, Ooka T, Itoh T, Sawaguchi A, Iguchi A, et al.. The Defective Prophage Pool of Escherichia coli O157: Prophage–Prophage Interactions Potentiate Horizontal Transfer of Virulence Determinants. PLoS Pathog. 2009. May 1;5(5):e1000408. doi: 10.1371/journal.ppat.1000408 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

