In diverse conditions, intrinsic chromatin condensates have liquid-like material properties
- PMID: 37094140
- PMCID: PMC10161002
- DOI: 10.1073/pnas.2218085120
In diverse conditions, intrinsic chromatin condensates have liquid-like material properties
Abstract
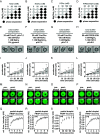
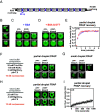
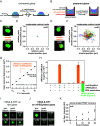
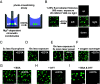
Nuclear DNA in eukaryotes is wrapped around histone proteins to form nucleosomes on a chromatin fiber. Dynamic folding of the chromatin fiber into loops and variations in the degree of chromatin compaction regulate essential processes such as transcription, recombination, and mitotic chromosome segregation. Our understanding of the physical properties that allow chromatin to be dynamically remodeled even in highly compacted states is limited. Previously, we reported that chromatin has an intrinsic capacity to phase separate and form dynamic liquid-like condensates, which can be regulated by cellular factors [B. A. Gibson et al., Cell 179, 470-484.e421 (2019)]. Recent contradictory reports claim that a specific set of solution conditions is required for fluidity in condensates that would otherwise be solid [J. C. Hansen, K. Maeshima, M. J. Hendzel, Epigenetics Chromatin 14, 50 (2021); H. Strickfaden et al., Cell 183, 1772-1784.e1713 (2020)]. We sought to resolve these discrepancies, as our ability to translate with confidence these biophysical observations to cells requires their precise characterization. Moreover, whether chromatin assemblies are dynamic or static affects how processes such as transcription, loop extrusion, and remodeling will engage them inside cells. Here, we show in diverse conditions and without specific buffering components that chromatin fragments form phase separated fluids in vitro. We also explore how sample preparation and imaging affect the experimental observation of chromatin condensate dynamics. Last, we describe how liquid-like in vitro behaviors can translate to the locally dynamic but globally constrained chromatin movement observed in cells.
Keywords: biomolecular condensate; chromatin; phase separation.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation.J Biol Chem. 2019 Feb 1;294(5):1451-1463. doi: 10.1074/jbc.RA118.006620. Epub 2018 Dec 4. J Biol Chem. 2019. PMID: 30514760 Free PMC article.
-
Mechanical properties of nucleoprotein complexes determined by nanoindentation spectroscopy.Nucleus. 2020 Dec;11(1):264-282. doi: 10.1080/19491034.2020.1816053. Nucleus. 2020. PMID: 32954931 Free PMC article.
-
HP1 reshapes nucleosome core to promote phase separation of heterochromatin.Nature. 2019 Nov;575(7782):390-394. doi: 10.1038/s41586-019-1669-2. Epub 2019 Oct 16. Nature. 2019. PMID: 31618757 Free PMC article.
-
Histone variant nucleosomes: structure, function and implication in disease.Subcell Biochem. 2007;41:71-89. Subcell Biochem. 2007. PMID: 17484124 Review.
-
Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology.Biochemistry. 2016 Mar 22;55(11):1584-99. doi: 10.1021/acs.biochem.5b01210. Epub 2016 Jan 26. Biochemistry. 2016. PMID: 26745824 Review.
Cited by
-
Chromatin accessibility: methods, mechanisms, and biological insights.Nucleus. 2022 Dec;13(1):236-276. doi: 10.1080/19491034.2022.2143106. Nucleus. 2022. PMID: 36404679 Free PMC article. Review.
-
Multiscale Bayesian simulations reveal functional chromatin condensation of gene loci.PNAS Nexus. 2024 Jun 6;3(6):pgae226. doi: 10.1093/pnasnexus/pgae226. eCollection 2024 Jun. PNAS Nexus. 2024. PMID: 38881841 Free PMC article.
-
Nucleosome spacing controls chromatin spatial structure and accessibility.Biophys J. 2024 Apr 2;123(7):847-857. doi: 10.1016/j.bpj.2024.02.024. Epub 2024 Feb 27. Biophys J. 2024. PMID: 38419332 Free PMC article.
-
Assessing the Effect of Chromatin-Binding Enzymes on Condensed Chromatin with Optical Tweezers.Methods Mol Biol. 2025;2881:345-355. doi: 10.1007/978-1-0716-4280-1_17. Methods Mol Biol. 2025. PMID: 39704952
-
ISWI catalyzes nucleosome sliding in condensed nucleosome arrays.Nat Struct Mol Biol. 2024 Sep;31(9):1331-1340. doi: 10.1038/s41594-024-01290-x. Epub 2024 Apr 25. Nat Struct Mol Biol. 2024. PMID: 38664566
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

