Allosteric communication between ligand binding domains modulates substrate inhibition in adenylate kinase
- PMID: 37094144
- PMCID: PMC10160949
- DOI: 10.1073/pnas.2219855120
Allosteric communication between ligand binding domains modulates substrate inhibition in adenylate kinase
Abstract
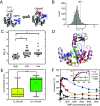
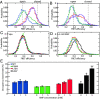
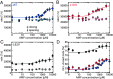
Enzymes play a vital role in life processes; they control chemical reactions and allow functional cycles to be synchronized. Many enzymes harness large-scale motions of their domains to achieve tremendous catalytic prowess and high selectivity for specific substrates. One outstanding example is provided by the three-domain enzyme adenylate kinase (AK), which catalyzes phosphotransfer between ATP to AMP. Here we study the phenomenon of substrate inhibition by AMP and its correlation with domain motions. Using single-molecule FRET spectroscopy, we show that AMP does not block access to the ATP binding site, neither by competitive binding to the ATP cognate site nor by directly closing the LID domain. Instead, inhibitory concentrations of AMP lead to a faster and more cooperative domain closure by ATP, leading in turn to an increased population of the closed state. The effect of AMP binding can be modulated through mutations throughout the structure of the enzyme, as shown by the screening of an extensive AK mutant library. The mutation of multiple conserved residues reduces substrate inhibition, suggesting that substrate inhibition is an evolutionary well conserved feature in AK. Combining these insights, we developed a model that explains the complex activity of AK, particularly substrate inhibition, based on the experimentally observed opening and closing rates. Notably, the model indicates that the catalytic power is affected by the microsecond balance between the open and closed states of the enzyme. Our findings highlight the crucial role of protein motions in enzymatic activity.
Keywords: enzymatic activity; protein dynamics; single-molecule FRET.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
Elucidating Dynamics of Adenylate Kinase from Enzyme Opening to Ligand Release.J Chem Inf Model. 2024 Jan 8;64(1):150-163. doi: 10.1021/acs.jcim.3c01618. Epub 2023 Dec 20. J Chem Inf Model. 2024. PMID: 38117131 Free PMC article.
-
Towards a mechanism of AMP-substrate inhibition in adenylate kinase from Escherichia coli.FEBS Lett. 1996 Nov 18;397(2-3):273-6. doi: 10.1016/s0014-5793(96)01195-7. FEBS Lett. 1996. PMID: 8955362
-
Substrate Binding Specifically Modulates Domain Arrangements in Adenylate Kinase.Biophys J. 2015 Nov 3;109(9):1978-85. doi: 10.1016/j.bpj.2015.08.049. Biophys J. 2015. PMID: 26536274 Free PMC article.
-
Folding funnels and conformational transitions via hinge-bending motions.Cell Biochem Biophys. 1999;31(2):141-64. doi: 10.1007/BF02738169. Cell Biochem Biophys. 1999. PMID: 10593256 Review.
-
Conformational heterogeneity within the LID domain mediates substrate binding to Escherichia coli adenylate kinase: function follows fluctuations.Top Curr Chem. 2013;337:95-121. doi: 10.1007/128_2012_410. Top Curr Chem. 2013. PMID: 23543318 Free PMC article. Review.
Cited by
-
Predicting binding events in very flexible, allosteric, multi-domain proteins.bioRxiv [Preprint]. 2024 Nov 10:2024.06.02.597018. doi: 10.1101/2024.06.02.597018. bioRxiv. 2024. Update in: J Chem Inf Model. 2025 Feb 24;65(4):2052-2065. doi: 10.1021/acs.jcim.4c01810. PMID: 38895346 Free PMC article. Updated. Preprint.
-
CDK2 and CDK4: Cell Cycle Functions Evolve Distinct, Catalysis-Competent Conformations, Offering Drug Targets.JACS Au. 2024 May 14;4(5):1911-1927. doi: 10.1021/jacsau.4c00138. eCollection 2024 May 27. JACS Au. 2024. PMID: 38818077 Free PMC article.
-
Interplay between conformational dynamics and substrate binding regulates enzymatic activity: a single-molecule FRET study.Chem Sci. 2025 Jan 22;16(7):3066-3077. doi: 10.1039/d4sc06819j. eCollection 2025 Feb 12. Chem Sci. 2025. PMID: 39877815 Free PMC article.
-
Enzyme activation by urea reveals the interplay between conformational dynamics and substrate binding: a single-molecule FRET study.bioRxiv [Preprint]. 2024 Sep 11:2024.09.01.610662. doi: 10.1101/2024.09.01.610662. bioRxiv. 2024. Update in: Chem Sci. 2025 Jan 22;16(7):3066-3077. doi: 10.1039/d4sc06819j. PMID: 39257823 Free PMC article. Updated. Preprint.
-
Allostery can convert binding free energies into concerted domain motions in enzymes.Nat Commun. 2024 Nov 22;15(1):10109. doi: 10.1038/s41467-024-54421-9. Nat Commun. 2024. PMID: 39572546 Free PMC article.
References
-
- Thirumalai D., Hyeon C., Zhuravlev P. I., Lorimer G. H., Symmetry, rigidity, and allosteric signaling: From monomeric proteins to molecular machines. Chem. Rev. 119, 6788–6821 (2019). - PubMed
-
- Henzler-Wildman K., Kern D., Dynamic personalities of proteins. Nature 450, 964–972 (2007). - PubMed
-
- Sinev M. A., Sineva E. V., Ittah V., Haas E., Domain closure in adenylate kinase. Biochemistry 35, 6425–6437 (1996). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

