HAPLN1 potentiates peritoneal metastasis in pancreatic cancer
- PMID: 37095087
- PMCID: PMC10126109
- DOI: 10.1038/s41467-023-38064-w
HAPLN1 potentiates peritoneal metastasis in pancreatic cancer
Abstract
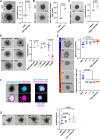
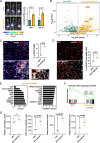
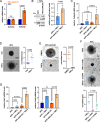
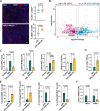
Pancreatic ductal adenocarcinoma (PDAC) frequently metastasizes into the peritoneum, which contributes to poor prognosis. Metastatic spreading is promoted by cancer cell plasticity, yet its regulation by the microenvironment is incompletely understood. Here, we show that the presence of hyaluronan and proteoglycan link protein-1 (HAPLN1) in the extracellular matrix enhances tumor cell plasticity and PDAC metastasis. Bioinformatic analysis showed that HAPLN1 expression is enriched in the basal PDAC subtype and associated with worse overall patient survival. In a mouse model for peritoneal carcinomatosis, HAPLN1-induced immunomodulation favors a more permissive microenvironment, which accelerates the peritoneal spread of tumor cells. Mechanistically, HAPLN1, via upregulation of tumor necrosis factor receptor 2 (TNFR2), promotes TNF-mediated upregulation of Hyaluronan (HA) production, facilitating EMT, stemness, invasion and immunomodulation. Extracellular HAPLN1 modifies cancer cells and fibroblasts, rendering them more immunomodulatory. As such, we identify HAPLN1 as a prognostic marker and as a driver for peritoneal metastasis in PDAC.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

