Post-transcriptional modification of m6A methylase METTL3 regulates ERK-induced androgen-deprived treatment resistance prostate cancer
- PMID: 37095108
- PMCID: PMC10126012
- DOI: 10.1038/s41419-023-05773-5
Post-transcriptional modification of m6A methylase METTL3 regulates ERK-induced androgen-deprived treatment resistance prostate cancer
Abstract
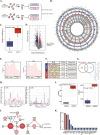
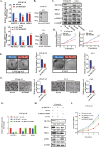
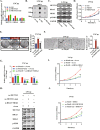
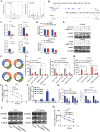
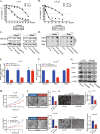
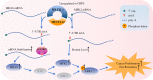
As the most common modification of RNA, N6-methyladenosin (m6A) has been confirmed to be involved in the occurrence and development of various cancers. However, the relationship between m6A and castration resistance prostate cancer (CRPC), has not been fully studied. By m6A-sequencing of patient cancer tissues, we identified that the overall level of m6A in CRPC was up-regulated than castration sensitive prostate cancer (CSPC). Based on the analysis of m6A-sequencing data, we found m6A modification level of HRas proto-oncogene, GTPase (HRAS) and mitogen-activated protein kinase kinase 2 (MEK2 or MAP2K2) were enhanced in CRPC. Specifically, tissue microarray analysis and molecular biology experiments confirmed that METTL3, an m6A "writer" up-regulated after castration, activated the ERK pathway to contribute to malignant phenotype including ADT resistance, cell proliferation and invasion. We revealed that METTL3-mediated ERK phosphorylation by stabilizing the transcription of HRAS and positively regulating the translation of MEK2. In the Enzalutamide-resistant (Enz-R) C4-2 and LNCap cell line (C4-2R, LNCapR) established in the current study, the ERK pathway was confirmed to be regulated by METTL3. We also found that applying antisense oligonucleotides (ASOs) to target the METTL3/ERK axis can restore Enzalutamide resistance in vitro and in vivo. In conclusion, METTL3 activated the ERK pathway and induced the resistance to Enzalutamide by regulating the m6A level of critical gene transcription in the ERK pathway.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests. All authors have made a significant contribution to the findings and methods described in the paper. All authors have reviewed and approved the manuscript. There are no financial or commercial interests, no ethical issues. The work has not been published and has not been submitted simultaneously to any other journal.
Figures


Similar articles
-
Aberrant activation of super enhancer and choline metabolism drive antiandrogen therapy resistance in prostate cancer.Oncogene. 2020 Oct;39(42):6556-6571. doi: 10.1038/s41388-020-01456-z. Epub 2020 Sep 11. Oncogene. 2020. PMID: 32917955
-
Targeting CPT1B as a potential therapeutic strategy in castration-resistant and enzalutamide-resistant prostate cancer.Prostate. 2020 Sep;80(12):950-961. doi: 10.1002/pros.24027. Epub 2020 Jul 10. Prostate. 2020. PMID: 32648618
-
Lycopene enhances the sensitivity of castration-resistant prostate cancer to enzalutamide through the AKT/EZH2/ androgen receptor signaling pathway.Biochem Biophys Res Commun. 2022 Jul 12;613:53-60. doi: 10.1016/j.bbrc.2022.04.126. Epub 2022 Apr 30. Biochem Biophys Res Commun. 2022. PMID: 35533600
-
Combination AZD5363 with Enzalutamide Significantly Delays Enzalutamide-resistant Prostate Cancer in Preclinical Models.Eur Urol. 2015 Jun;67(6):986-990. doi: 10.1016/j.eururo.2014.08.006. Epub 2014 Aug 20. Eur Urol. 2015. PMID: 25151012
-
Natural product myricetin is a pan-KDM4 inhibitor which with poly lactic-co-glycolic acid formulation effectively targets castration-resistant prostate cancer.J Biomed Sci. 2022 May 9;29(1):29. doi: 10.1186/s12929-022-00812-3. J Biomed Sci. 2022. PMID: 35534851 Free PMC article.
Cited by
-
Icariin-Curcumol promotes docetaxel sensitivity in prostate cancer through modulation of the PI3K-Akt signaling pathway and the Warburg effect.Cancer Cell Int. 2023 Sep 2;23(1):190. doi: 10.1186/s12935-023-03042-1. Cancer Cell Int. 2023. PMID: 37660001 Free PMC article.
-
The current landscape of m6A modification in urological cancers.PeerJ. 2023 Sep 7;11:e16023. doi: 10.7717/peerj.16023. eCollection 2023. PeerJ. 2023. PMID: 37701836 Free PMC article. Review.
-
FMNL2/SRC-mediated androgen receptor translocation into the nucleus promotes enzalutamide resistance of prostate cancer.iScience. 2025 Mar 12;28(4):112205. doi: 10.1016/j.isci.2025.112205. eCollection 2025 Apr 18. iScience. 2025. PMID: 40212590 Free PMC article.
-
Immunomodulatory role of RNA modifications in sex hormone-dependent cancers.Front Immunol. 2025 May 8;16:1513037. doi: 10.3389/fimmu.2025.1513037. eCollection 2025. Front Immunol. 2025. PMID: 40406121 Free PMC article. Review.
-
ELOVL2 mediated stabilization of AR contributes to enzalutamide resistance in prostate cancer.Front Cell Dev Biol. 2025 Jun 9;13:1598400. doi: 10.3389/fcell.2025.1598400. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40552308 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

