TMEM25 inhibits monomeric EGFR-mediated STAT3 activation in basal state to suppress triple-negative breast cancer progression
- PMID: 37095176
- PMCID: PMC10126118
- DOI: 10.1038/s41467-023-38115-2
TMEM25 inhibits monomeric EGFR-mediated STAT3 activation in basal state to suppress triple-negative breast cancer progression
Abstract
Triple-negative breast cancer (TNBC) is a subtype of breast cancer with poor outcome and lacks of approved targeted therapy. Overexpression of epidermal growth factor receptor (EGFR) is found in more than 50% TNBC and is suggested as a driving force in progression of TNBC; however, targeting EGFR using antibodies to prevent its dimerization and activation shows no significant benefits for TNBC patients. Here we report that EGFR monomer may activate signal transducer activator of transcription-3 (STAT3) in the absence of transmembrane protein TMEM25, whose expression is frequently decreased in human TNBC. Deficiency of TMEM25 allows EGFR monomer to phosphorylate STAT3 independent of ligand binding, and thus enhances basal STAT3 activation to promote TNBC progression in female mice. Moreover, supplying TMEM25 by adeno-associated virus strongly suppresses STAT3 activation and TNBC progression. Hence, our study reveals a role of monomeric-EGFR/STAT3 signaling pathway in TNBC progression and points out a potential targeted therapy for TNBC.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
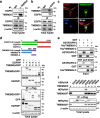
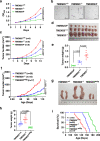
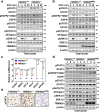


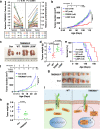
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

