Characterization of the m6A regulator-mediated methylation modification patterns in oral squamous cell carcinoma
- PMID: 37095314
- PMCID: PMC10126108
- DOI: 10.1038/s41598-023-33891-9
Characterization of the m6A regulator-mediated methylation modification patterns in oral squamous cell carcinoma
Abstract
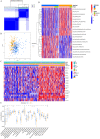
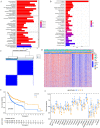
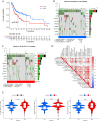
N6-methyladenosine (m6A) is a form of posttranscriptional modification that plays important roles in cancer including oral squamous cell carcinoma (OSCC). Most studies to date have focused on a limited number of regulators and oncogenic pathways, thus failing to provide comprehensive insight into the dynamic effects of m6A modification. In addition, the role of m6A modification in shaping immune cell infiltration in OSCC has yet to be clarified. This study was designed to assess m6A modification dynamics in OSCC and to understand how such modifications influence clinical immunotherapeutic treatment outcomes. m6A modification patterns linked with 23 m6A regulators were analyzed in 437 OSCC patients from TCGA and GEO cohorts. These patterns were then quantified through m6A score based on algorithms derived from a principal component analysis (PCA) approach. The m6A modification patterns of OSCC samples were grouped into two clusters based on the m6A regulators expression, and immune cell infiltration was linked with the 5-year survival outcomes of patients in these clusters. 1575 genes associated with OSCC patient prognosis were identified and used to re-cluster these samples into two groups. Patients in clusters exhibiting higher levels of m6A regulator expression exhibited poorer overall survival (OS), whereas patients with high m6A scores survived for longer (p < 0.001). The overall mortality rates in the groups of patients with low and high m6A scores were 55% and 40%, respectively, and the m6A score distributions in clusters of patients grouped by m6A modification patterns and gene expression further supported the link between a high m6A score and better prognostic outcomes. Immunophenoscore (IPS) values for patients in different m6A score groups suggested that the use of PD-1-specific antibodies or CTLA-4 inhibitors alone or in combination would yield superior treatment outcomes in patients in the high-m6A score group relative to the low-m6A score group. m6A modification patterns are relevant to heterogeneity in OSCC. Detailed analyses of m6A modification patterns may thus offer novel insight regarding immune cell infiltration within the OSCC tumor microenvironment, guiding novel efforts to provide patients with more effective immunotherapeutic interventions.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
A Novel Immune-Related Gene Signature to Identify the Tumor Microenvironment and Prognose Disease Among Patients With Oral Squamous Cell Carcinoma Patients Using ssGSEA: A Bioinformatics and Biological Validation Study.Front Immunol. 2022 Jul 6;13:922195. doi: 10.3389/fimmu.2022.922195. eCollection 2022. Front Immunol. 2022. PMID: 35935989 Free PMC article.
-
The Impact of m1A Methylation Modification Patterns on Tumor Immune Microenvironment and Prognosis in Oral Squamous Cell Carcinoma.Int J Mol Sci. 2021 Sep 24;22(19):10302. doi: 10.3390/ijms221910302. Int J Mol Sci. 2021. PMID: 34638642 Free PMC article.
-
A novel T-cell proliferation-associated gene predicts prognosis and reveals immune infiltration in patients with oral squamous cell carcinoma.Arch Oral Biol. 2023 Aug;152:105719. doi: 10.1016/j.archoralbio.2023.105719. Epub 2023 May 9. Arch Oral Biol. 2023. PMID: 37178584
-
Tumor-infiltrating ICOS+ Effector Regulatory T-Cells in Oral Squamous Cell Carcinoma as a Promising Biomarker for Prognosis and 'Hot' Tumor.Anticancer Res. 2022 May;42(5):2383-2393. doi: 10.21873/anticanres.15717. Anticancer Res. 2022. PMID: 35489733 Review.
-
Oral squamous cell carcinomas: state of the field and emerging directions.Int J Oral Sci. 2023 Sep 22;15(1):44. doi: 10.1038/s41368-023-00249-w. Int J Oral Sci. 2023. PMID: 37736748 Free PMC article. Review.
Cited by
-
RNA modifications in cancer immune therapy: regulators of immune cells and immune checkpoints.Front Immunol. 2024 Sep 20;15:1463847. doi: 10.3389/fimmu.2024.1463847. eCollection 2024. Front Immunol. 2024. PMID: 39372415 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

