High-dimensional analysis of 16 SARS-CoV-2 vaccine combinations reveals lymphocyte signatures correlating with immunogenicity
- PMID: 37095378
- PMCID: PMC10232362
- DOI: 10.1038/s41590-023-01499-w
High-dimensional analysis of 16 SARS-CoV-2 vaccine combinations reveals lymphocyte signatures correlating with immunogenicity
Abstract
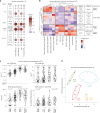
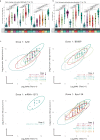
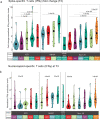
The range of vaccines developed against severe acute respiratory syndrome coronavirus 2 (SARS‑CoV‑2) provides a unique opportunity to study immunization across different platforms. In a single-center cohort, we analyzed the humoral and cellular immune compartments following five coronavirus disease 2019 (COVID-19) vaccines spanning three technologies (adenoviral, mRNA and inactivated virus) administered in 16 combinations. For adenoviral and inactivated-virus vaccines, heterologous combinations were generally more immunogenic compared to homologous regimens. The mRNA vaccine as the second dose resulted in the strongest antibody response and induced the highest frequency of spike-binding memory B cells irrespective of the priming vaccine. Priming with the inactivated-virus vaccine increased the SARS-CoV-2-specific T cell response, whereas boosting did not. Distinct immune signatures were elicited by the different vaccine combinations, demonstrating that the immune response is shaped by the type of vaccines applied and the order in which they are delivered. These data provide a framework for improving future vaccine strategies against pathogens and cancer.
Trial registration: ClinicalTrials.gov NCT04988048.
© 2023. The Author(s).
Conflict of interest statement
All authors declare no competing interests.
Figures











References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

