Candidate variants in DNA replication and repair genes in early-onset renal cell carcinoma patients referred for germline testing
- PMID: 37095444
- PMCID: PMC10123997
- DOI: 10.1186/s12864-023-09310-8
Candidate variants in DNA replication and repair genes in early-onset renal cell carcinoma patients referred for germline testing
Erratum in
-
Correction: Candidate variants in DNA replication and repair genes in early-onset renal cell carcinoma patients referred for germline testing.BMC Genomics. 2023 Jul 10;24(1):388. doi: 10.1186/s12864-023-09486-z. BMC Genomics. 2023. PMID: 37430226 Free PMC article. No abstract available.
Abstract
Background: Early-onset renal cell carcinoma (eoRCC) is typically associated with pathogenic germline variants (PGVs) in RCC familial syndrome genes. However, most eoRCC patients lack PGVs in familial RCC genes and their genetic risk remains undefined.
Methods: Here, we analyzed biospecimens from 22 eoRCC patients that were seen at our institution for genetic counseling and tested negative for PGVs in RCC familial syndrome genes.
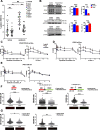
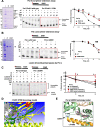
Results: Analysis of whole-exome sequencing (WES) data found enrichment of candidate pathogenic germline variants in DNA repair and replication genes, including multiple DNA polymerases. Induction of DNA damage in peripheral blood monocytes (PBMCs) significantly elevated numbers of [Formula: see text]H2AX foci, a marker of double-stranded breaks, in PBMCs from eoRCC patients versus PBMCs from matched cancer-free controls. Knockdown of candidate variant genes in Caki RCC cells increased [Formula: see text]H2AX foci. Immortalized patient-derived B cell lines bearing the candidate variants in DNA polymerase genes (POLD1, POLH, POLE, POLK) had DNA replication defects compared to control cells. Renal tumors carrying these DNA polymerase variants were microsatellite stable but had a high mutational burden. Direct biochemical analysis of the variant Pol δ and Pol η polymerases revealed defective enzymatic activities.
Conclusions: Together, these results suggest that constitutional defects in DNA repair underlie a subset of eoRCC cases. Screening patient lymphocytes to identify these defects may provide insight into mechanisms of carcinogenesis in a subset of genetically undefined eoRCCs. Evaluation of DNA repair defects may also provide insight into the cancer initiation mechanisms for subsets of eoRCCs and lay the foundation for targeting DNA repair vulnerabilities in eoRCC.
Keywords: DNA repair; DNA replication; Germline; Renal cancer.
© 2023. The Author(s).
Conflict of interest statement
All authors declare that there is no competing interest regarding this manuscript. M.J.H. performs collaborative research (with no funding) with the following: Myriad Genetics, Invitae Corporation, Ambry Genetics, Foundation Medicine, Inc. He also performs collaborative research (with no funding) and is part of a Precision Oncology Alliance funded by Caris Life Sciences (cover travel and meals at meetings). S.A. performs collaborative research (with no funding) with Caris Life Sciences, Foundation Medicine, Inc., Ambry Genetics, and Invitae Corporation. S.A.’s spouse is employed by Akoya Biosciences and has stocks in Akoya Biosciences, HTG Molecular Diagnostics, Abcam Plc., and Senzo Health. S.A., M.J.H., E.A.G., I.G.S. have patents and/or pending patents related to cancer diagnostics/treatment.
Figures



References
-
- Rosner G, Gluck N, Carmi S, Bercovich D, Fliss-Issakov N, Ben-Yehoyada M, Aharon-Caspi S, Kellerman E, Strul H, Shibolet O, et al. POLD1 and POLE gene mutations in Jewish cohorts of early-onset colorectal cancer and of multiple colorectal adenomas. Dis Colon Rectum. 2018;61(9):1073–1079. doi: 10.1097/DCR.0000000000001150. - DOI - PubMed
-
- Carlo MI, Mukherjee S, Mandelker D, Vijai J, Kemel Y, Zhang L, Knezevic A, Patil S, Ceyhan-Birsoy O, Huang KC, et al. Prevalence of germline mutations in cancer susceptibility genes in patients with advanced renal cell carcinoma. JAMA Oncol. 2018;4(9):1228–1235. doi: 10.1001/jamaoncol.2018.1986. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

