Untargeted lipidomic analysis and network pharmacology for parthenolide treated papillary thyroid carcinoma cells
- PMID: 37095470
- PMCID: PMC10123985
- DOI: 10.1186/s12906-023-03944-7
Untargeted lipidomic analysis and network pharmacology for parthenolide treated papillary thyroid carcinoma cells
Abstract
Background: With fast rising incidence, papillary thyroid carcinoma (PTC) is the most common head and neck cancer. Parthenolide, isolated from traditional Chinese medicine, inhibits various cancer cells, including PTC cells. The aim was to investigate the lipid profile and lipid changes of PTC cells when treated with parthenolide.
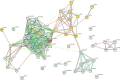
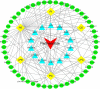
Methods: Comprehensive lipidomic analysis of parthenolide treated PTC cells was conducted using a UHPLC/Q-TOF-MS platform, and the changed lipid profile and specific altered lipid species were explored. Network pharmacology and molecular docking were performed to show the associations among parthenolide, changed lipid species, and potential target genes.
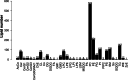
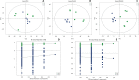
Results: With high stability and reproducibility, a total of 34 lipid classes and 1736 lipid species were identified. Lipid class analysis indicated that parthenolide treated PTC cells contained higher levels of fatty acid (FA), cholesterol ester (ChE), simple glc series 3 (CerG3) and lysophosphatidylglycerol (LPG), lower levels of zymosterol (ZyE) and Monogalactosyldiacylglycerol (MGDG) than controlled ones, but with no significant differences. Several specific lipid species were changed significantly in PTC cells treated by parthenolide, including the increasing of phosphatidylcholine (PC) (12:0e/16:0), PC (18:0/20:4), CerG3 (d18:1/24:1), lysophosphatidylethanolamine (LPE) (18:0), phosphatidylinositol (PI) (19:0/20:4), lysophosphatidylcholine (LPC) (28:0), ChE (22:6), and the decreasing of phosphatidylethanolamine (PE) (16:1/17:0), PC (34:1) and PC (16:0p/18:0). Four key targets (PLA2G4A, LCAT, LRAT, and PLA2G2A) were discovered when combining network pharmacology and lipidomics. Among them, PLA2G2A and PLA2G4A were able to bind with parthenolide confirmed by molecular docking.
Conclusions: The changed lipid profile and several significantly altered lipid species of parthenolide treated PTC cells were observed. These altered lipid species, such as PC (34:1), and PC (16:0p/18:0), may be involved in the antitumor mechanisms of parthenolide. PLA2G2A and PLA2G4A may play key roles when parthenolide treated PTC cells.
Keywords: Lipidomics; Network pharmacology; Parthenolide; Phosphatidylcholine; Thyroid cancer.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
[Nontargeted lipidomic analysis of sera from sepsis patients based on ultra-high performance liquid chromatography-mass spectrometry/mass spectrometry].Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2022 Apr;34(4):346-351. doi: 10.3760/cma.j.cn121430-20210612-00875. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2022. PMID: 35692196 Chinese.
-
Development of lipidomic platform and phosphatidylcholine retention time index for lipid profiling of rosuvastatin treated human plasma.J Chromatogr B Analyt Technol Biomed Life Sci. 2014 Jan 1;944:157-65. doi: 10.1016/j.jchromb.2013.10.029. Epub 2013 Oct 26. J Chromatogr B Analyt Technol Biomed Life Sci. 2014. PMID: 24316528
-
Plasma lipidomics reveals potential lipid markers of major depressive disorder.Anal Bioanal Chem. 2016 Sep;408(23):6497-507. doi: 10.1007/s00216-016-9768-5. Epub 2016 Jul 25. Anal Bioanal Chem. 2016. PMID: 27457104
-
Combined metabolomic and lipidomic analysis uncovers metabolic profile and biomarkers for papillary thyroid carcinoma.Sci Rep. 2023 Oct 17;13(1):17666. doi: 10.1038/s41598-023-41176-4. Sci Rep. 2023. PMID: 37848492 Free PMC article.
-
Lipidomics as a principal tool for advancing biomedical research.J Genet Genomics. 2013 Aug 20;40(8):375-90. doi: 10.1016/j.jgg.2013.06.007. Epub 2013 Jul 12. J Genet Genomics. 2013. PMID: 23969247 Review.
Cited by
-
The genetic associations of lipidome on bladder cancer: a Mendelian randomization study.Discov Oncol. 2025 Apr 22;16(1):586. doi: 10.1007/s12672-025-02389-3. Discov Oncol. 2025. PMID: 40261590 Free PMC article.
-
The Role of Lipid Metabolism Disorders in the Development of Thyroid Cancer.Int J Mol Sci. 2024 Jun 28;25(13):7129. doi: 10.3390/ijms25137129. Int J Mol Sci. 2024. PMID: 39000236 Free PMC article. Review.
-
Network pharmacology: an efficient but underutilized approach in oral, head and neck cancer therapy-a review.Front Pharmacol. 2024 Jul 5;15:1410942. doi: 10.3389/fphar.2024.1410942. eCollection 2024. Front Pharmacol. 2024. PMID: 39035991 Free PMC article. Review.
References
-
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. - PubMed
-
- Wang X, Lu X, Geng Z, Yang G, Shi Y. LncRNA PTCSC3/miR-574-5p governs cell proliferation and migration of papillary thyroid carcinoma via Wnt/β-Catenin signaling. J Cell Biochem. 2017;118(12):4745–4752. - PubMed
-
- Sipos JA, Mazzaferri EL. Thyroid cancer epidemiology and prognostic variables. Clin Oncol (R Coll Radiol) 2010;22(6):395–404. - PubMed
-
- Gao W, Li L, Zhang X, et al. Nanomagnetic liposome-encapsulated parthenolide and indocyanine green for targeting and chemo-photothermal antitumor therapy. Nanomedicine (Lond) 2020;15(9):871–890. - PubMed
-
- Yuan L, Wang Z, Zhang D, Wang J. Metabonomic study of the intervention effects of Parthenolide on anti-thyroid cancer activity. J Chromatogr B Analyt Technol Biomed Life Sci. 2020;1150:122179. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

