A desert lncRNA HIDEN regulates human endoderm differentiation via interacting with IMP1 and stabilizing FZD5 mRNA
- PMID: 37095549
- PMCID: PMC10124006
- DOI: 10.1186/s13059-023-02925-w
A desert lncRNA HIDEN regulates human endoderm differentiation via interacting with IMP1 and stabilizing FZD5 mRNA
Abstract
Background: Extensive studies have revealed the function and mechanism of lncRNAs in development and differentiation, but the majority have focused on those lncRNAs adjacent to protein-coding genes. In contrast, lncRNAs located in gene deserts are rarely explored. Here, we utilize multiple differentiation systems to dissect the role of a desert lncRNA, HIDEN (human IMP1-associated "desert" definitive endoderm lncRNA), in definitive endoderm differentiation from human pluripotent stem cells.
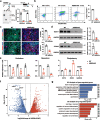
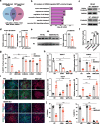
Results: We show that desert lncRNAs are highly expressed with cell-stage-specific patterns and conserved subcellular localization during stem cell differentiation. We then focus on the desert lncRNA HIDEN which is upregulated and plays a vital role during human endoderm differentiation. We find depletion of HIDEN by either shRNA or promoter deletion significantly impairs human endoderm differentiation. HIDEN functionally interacts with RNA-binding protein IMP1 (IGF2BP1), which is also required for endoderm differentiation. Loss of HIDEN or IMP1 results in reduced WNT activity, and WNT agonist rescues endoderm differentiation deficiency caused by the depletion of HIDEN or IMP1. Moreover, HIDEN depletion reduces the interaction between IMP1 protein and FZD5 mRNA and causes the destabilization of FZD5 mRNA, which is a WNT receptor and necessary for definitive endoderm differentiation.
Conclusions: These data suggest that desert lncRNA HIDEN facilitates the interaction between IMP1 and FZD5 mRNA, stabilizing FZD5 mRNA which activates WNT signaling and promotes human definitive endoderm differentiation.
Keywords: Desert lncRNA; Endoderm differentiation; FZD5; Human pluripotent stem cell; IMP1.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
GATA6-AS1 Regulates GATA6 Expression to Modulate Human Endoderm Differentiation.Stem Cell Reports. 2020 Sep 8;15(3):694-705. doi: 10.1016/j.stemcr.2020.07.014. Epub 2020 Aug 13. Stem Cell Reports. 2020. PMID: 32795420 Free PMC article.
-
DIGIT Is a Conserved Long Noncoding RNA that Regulates GSC Expression to Control Definitive Endoderm Differentiation of Embryonic Stem Cells.Cell Rep. 2016 Oct 4;17(2):353-365. doi: 10.1016/j.celrep.2016.09.017. Cell Rep. 2016. PMID: 27705785 Free PMC article.
-
The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression.Cell Rep. 2015 Apr 7;11(1):137-48. doi: 10.1016/j.celrep.2015.03.008. Epub 2015 Apr 2. Cell Rep. 2015. PMID: 25843708 Free PMC article.
-
Long noncoding RNA as an emerging regulator of endoderm differentiation: progress and perspectives.Cell Regen. 2025 Mar 26;14(1):11. doi: 10.1186/s13619-025-00230-4. Cell Regen. 2025. PMID: 40133743 Free PMC article. Review.
-
Long Noncoding RNAs in Human Stemness and Differentiation.Trends Cell Biol. 2021 Jul;31(7):542-555. doi: 10.1016/j.tcb.2021.02.002. Epub 2021 Mar 1. Trends Cell Biol. 2021. PMID: 33663944 Review.
Cited by
-
Wnt/β-catenin signaling pathway: proteins' roles in osteoporosis and cancer diseases and the regulatory effects of natural compounds on osteoporosis.Mol Med. 2024 Oct 28;30(1):193. doi: 10.1186/s10020-024-00957-x. Mol Med. 2024. PMID: 39468464 Free PMC article. Review.
-
The Role of Long Non-Coding RNAs in Human Endoderm Differentiation.Noncoding RNA. 2025 Apr 13;11(2):29. doi: 10.3390/ncrna11020029. Noncoding RNA. 2025. PMID: 40278506 Free PMC article. Review.
-
Molecular subgroup establishment and signature creation of lncRNAs associated with acetylation in lung adenocarcinoma.Aging (Albany NY). 2024 Jan 17;16(2):1276-1297. doi: 10.18632/aging.205407. Epub 2024 Jan 17. Aging (Albany NY). 2024. PMID: 38240708 Free PMC article.
-
Prostatic lineage differentiation from human embryonic stem cells through inducible expression of NKX3-1.Stem Cell Res Ther. 2024 Sep 2;15(1):274. doi: 10.1186/s13287-024-03886-y. Stem Cell Res Ther. 2024. PMID: 39218930 Free PMC article.
-
A noncoding variant confers pancreatic differentiation defect and contributes to diabetes susceptibility by recruiting RXRA.Nat Commun. 2024 Nov 12;15(1):9771. doi: 10.1038/s41467-024-54151-y. Nat Commun. 2024. PMID: 39532884 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

