Combining different CRISPR nucleases for simultaneous knock-in and base editing prevents translocations in multiplex-edited CAR T cells
- PMID: 37095570
- PMCID: PMC10123993
- DOI: 10.1186/s13059-023-02928-7
Combining different CRISPR nucleases for simultaneous knock-in and base editing prevents translocations in multiplex-edited CAR T cells
Erratum in
-
Author Correction: Combining different CRISPR nucleases for simultaneous knock-in and base editing prevents translocations in multiplex-edited CAR T cells.Genome Biol. 2025 Mar 26;26(1):71. doi: 10.1186/s13059-025-03548-z. Genome Biol. 2025. PMID: 40140907 Free PMC article. No abstract available.
Abstract
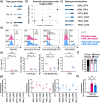
Background: Multiple genetic modifications may be required to develop potent off-the-shelf chimeric antigen receptor (CAR) T cell therapies. Conventional CRISPR-Cas nucleases install sequence-specific DNA double-strand breaks (DSBs), enabling gene knock-out or targeted transgene knock-in. However, simultaneous DSBs provoke a high rate of genomic rearrangements which may impede the safety of the edited cells.
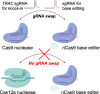
Results: Here, we combine a non-viral CRISPR-Cas9 nuclease-assisted knock-in and Cas9-derived base editing technology for DSB free knock-outs within a single intervention. We demonstrate efficient insertion of a CAR into the T cell receptor alpha constant (TRAC) gene, along with two knock-outs that silence major histocompatibility complexes (MHC) class I and II expression. This approach reduces translocations to 1.4% of edited cells. Small insertions and deletions at the base editing target sites indicate guide RNA exchange between the editors. This is overcome by using CRISPR enzymes of distinct evolutionary origins. Combining Cas12a Ultra for CAR knock-in and a Cas9-derived base editor enables the efficient generation of triple-edited CAR T cells with a translocation frequency comparable to unedited T cells. Resulting TCR- and MHC-negative CAR T cells resist allogeneic T cell targeting in vitro.
Conclusions: We outline a solution for non-viral CAR gene transfer and efficient gene silencing using different CRISPR enzymes for knock-in and base editing to prevent translocations. This single-step procedure may enable safer multiplex-edited cell products and demonstrates a path towards off-the-shelf CAR therapeutics.
© 2023. The Author(s).
Conflict of interest statement
Charité has received reagents for gene editing (
Figures




Similar articles
-
Quadruple adenine base-edited allogeneic CAR T cells outperform CRISPR/Cas9 nuclease-engineered T cells.Proc Natl Acad Sci U S A. 2025 May 20;122(20):e2427216122. doi: 10.1073/pnas.2427216122. Epub 2025 May 5. Proc Natl Acad Sci U S A. 2025. PMID: 40324075
-
An aptamer-mediated base editing platform for simultaneous knockin and multiple gene knockout for allogeneic CAR-T cells generation.Mol Ther. 2024 Aug 7;32(8):2692-2710. doi: 10.1016/j.ymthe.2024.06.033. Epub 2024 Jun 26. Mol Ther. 2024. PMID: 38937969 Free PMC article.
-
Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors.Nat Commun. 2019 Nov 19;10(1):5222. doi: 10.1038/s41467-019-13007-6. Nat Commun. 2019. PMID: 31745080 Free PMC article.
-
Review: Sustainable Clinical Development of CAR-T Cells - Switching From Viral Transduction Towards CRISPR-Cas Gene Editing.Front Immunol. 2022 Jun 17;13:865424. doi: 10.3389/fimmu.2022.865424. eCollection 2022. Front Immunol. 2022. PMID: 35784280 Free PMC article. Review.
-
INDEL detection, the 'Achilles heel' of precise genome editing: a survey of methods for accurate profiling of gene editing induced indels.Nucleic Acids Res. 2020 Dec 2;48(21):11958-11981. doi: 10.1093/nar/gkaa975. Nucleic Acids Res. 2020. PMID: 33170255 Free PMC article. Review.
Cited by
-
Mapping variant effects on anti-tumor hallmarks of primary human T cells with base-editing screens.Nat Biotechnol. 2025 Mar;43(3):384-395. doi: 10.1038/s41587-024-02235-x. Epub 2024 May 23. Nat Biotechnol. 2025. PMID: 38783148
-
Applications of CRISPR technology in cellular immunotherapy.Immunol Rev. 2023 Nov;320(1):199-216. doi: 10.1111/imr.13241. Epub 2023 Jul 14. Immunol Rev. 2023. PMID: 37449673 Free PMC article. Review.
-
Precise, predictable genome integrations by deep-learning-assisted design of microhomology-based templates.Nat Biotechnol. 2025 Aug 12. doi: 10.1038/s41587-025-02771-0. Online ahead of print. Nat Biotechnol. 2025. PMID: 40796977
-
Recent updates on allogeneic CAR-T cells in hematological malignancies.Cancer Cell Int. 2024 Sep 3;24(1):304. doi: 10.1186/s12935-024-03479-y. Cancer Cell Int. 2024. PMID: 39227937 Free PMC article. Review.
-
Integration of ζ-deficient CARs into the CD3ζ gene conveys potent cytotoxicity in T and NK cells.Blood. 2024 Jun 20;143(25):2599-2611. doi: 10.1182/blood.2023020973. Blood. 2024. PMID: 38493479 Free PMC article.
References
-
- Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat Rev Drug Discovery. 2020;19(3):185–99. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

