A myeloid leukemia factor homolog is involved in tolerance to stresses and stress-induced protein metabolism in Giardia lamblia
- PMID: 37095576
- PMCID: PMC10127389
- DOI: 10.1186/s13062-023-00378-6
A myeloid leukemia factor homolog is involved in tolerance to stresses and stress-induced protein metabolism in Giardia lamblia
Abstract
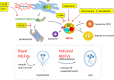
Background: The eukaryotic membrane vesicles contain specific sets of proteins that determine vesicle function and shuttle with specific destination. Giardia lamblia contains unknown cytosolic vesicles that are related to the identification of a homolog of human myeloid leukemia factor (MLF) named MLF vesicles (MLFVs). Previous studies suggest that MLF also colocalized with two autophagy machineries, FYVE and ATG8-like protein, and that MLFVs are stress-induced compartments for substrates of the proteasome or autophagy in response to rapamycin, MG132, and chloroquine treatment. A mutant protein of cyclin-dependent kinase 2, CDK2m3, was used to understand whether the aberrant proteins are targeted to degradative compratments. Interestingly, MLF was upregulated by CDK2m3 and they both colocalized within the same vesicles. Autophagy is a self-digestion process that is activated to remove damaged proteins for preventing cell death in response to various stresses. Because of the absence of some autophagy machineries, the mechanism of autophagy is unclear in G. lamblia.
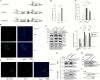
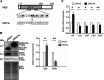
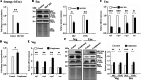
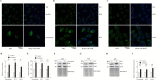
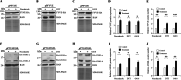
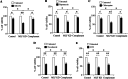
Results: In this study, we tested the six autophagosome and stress inducers in mammalian cells, including MG132, rapamycin, chloroquine, nocodazole, DTT, and G418, and found that their treatment increased reactive oxygen species production and vesicle number and level of MLF, FYVE, and ATG8-like protein in G. lamblia. Five stress inducers also increased the CDK2m3 protein levels and vesicles. Using stress inducers and knockdown system for MLF, we identified that stress induction of CDK2m3 was positively regulated by MLF. An autophagosome-reducing agent, 3-methyl adenine, can reduce MLF and CDK2m3 vesicles and proteins. In addition, knockdown of MLF with CRISPR/Cas9 system reduced cell survival upon treatment with stress inducers. Our newly developed complementation system for CRISPR/Cas9 indicated that complementation of MLF restored cell survival in response to stress inducers. Furthermore, human MLF2, like Giardia MLF, can increase cyst wall protein expression and cyst formation in G. lamblia, and it can colocalize with MLFVs and interact with MLF.
Conclusions: Our results suggest that MLF family proteins are functionally conserved in evolution. Our results also suggest an important role of MLF in survival in stress conditions and that MLFVs share similar stress-induced characteristics with autophagy compartments.
Keywords: ATG8-like; CDK2 mutant; Cyst; DTT; Differentiation; FYVE; G418; Giardia; MLF; Nocodazole.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
A myeloid leukemia factor homolog involved in encystation-induced protein metabolism in Giardia lamblia.Biochim Biophys Acta Gen Subj. 2021 Jun;1865(6):129859. doi: 10.1016/j.bbagen.2021.129859. Epub 2021 Feb 11. Biochim Biophys Acta Gen Subj. 2021. PMID: 33581251
-
Development of CRISPR/Cas9-mediated gene disruption systems in Giardia lamblia.PLoS One. 2019 Mar 11;14(3):e0213594. doi: 10.1371/journal.pone.0213594. eCollection 2019. PLoS One. 2019. PMID: 30856211 Free PMC article.
-
Identification of target genes regulated by encystation-induced transcription factor Myb2 using knockout mutagenesis in Giardia lamblia.Parasit Vectors. 2022 Oct 7;15(1):360. doi: 10.1186/s13071-022-05489-z. Parasit Vectors. 2022. PMID: 36207732 Free PMC article.
-
Protein trafficking in Giardia lamblia.Cell Microbiol. 2003 Jul;5(7):427-34. doi: 10.1046/j.1462-5822.2003.00284.x. Cell Microbiol. 2003. PMID: 12814433 Review.
-
Mechanisms of adaptation in the intestinal parasite Giardia lamblia.Essays Biochem. 2011;51:177-91. doi: 10.1042/bse0510177. Essays Biochem. 2011. PMID: 22023449 Review.
Cited by
-
Mlf mediates proteotoxic response via formation of cellular foci for protein folding and degradation in Giardia.PLoS Pathog. 2024 Oct 21;20(10):e1012617. doi: 10.1371/journal.ppat.1012617. eCollection 2024 Oct. PLoS Pathog. 2024. PMID: 39432513 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

