Nucleic acid strand displacement - from DNA nanotechnology to translational regulation
- PMID: 37095744
- PMCID: PMC10132225
- DOI: 10.1080/15476286.2023.2204565
Nucleic acid strand displacement - from DNA nanotechnology to translational regulation
Abstract
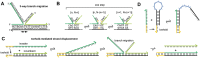
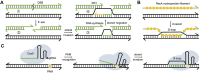
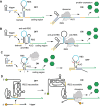
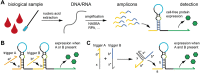
Nucleic acid strand displacement reactions involve the competition of two or more DNA or RNA strands of similar sequence for binding to a complementary strand, and facilitate the isothermal replacement of an incumbent strand by an invader. The process can be biased by augmenting the duplex comprising the incumbent with a single-stranded extension, which can act as a toehold for a complementary invader. The toehold gives the invader a thermodynamic advantage over the incumbent, and can be programmed as a unique label to activate a specific strand displacement process. Toehold-mediated strand displacement processes have been extensively utilized for the operation of DNA-based molecular machines and devices as well as for the design of DNA-based chemical reaction networks. More recently, principles developed initially in the context of DNA nanotechnology have been applied for the de novo design of gene regulatory switches that can operate inside living cells. The article specifically focuses on the design of RNA-based translational regulators termed toehold switches. Toehold switches utilize toehold-mediated strand invasion to either activate or repress translation of an mRNA in response to the binding of a trigger RNA molecule. The basic operation principles of toehold switches will be discussed as well as their applications in sensing and biocomputing. Finally, strategies for their optimization will be described as well as challenges for their operation in vivo.
Keywords: RNA synthetic biology; Strand displacement; riboregulators.
Conflict of interest statement
No potential conflict of interest was reported by the author.
Figures
Similar articles
-
Genetic switches based on nucleic acid strand displacement.Curr Opin Biotechnol. 2023 Feb;79:102867. doi: 10.1016/j.copbio.2022.102867. Epub 2022 Dec 17. Curr Opin Biotechnol. 2023. PMID: 36535150 Review.
-
Kinetics of RNA and RNA:DNA Hybrid Strand Displacement.ACS Synth Biol. 2021 Nov 19;10(11):3066-3073. doi: 10.1021/acssynbio.1c00336. Epub 2021 Nov 9. ACS Synth Biol. 2021. PMID: 34752075
-
Principles and Applications of Nucleic Acid Strand Displacement Reactions.Chem Rev. 2019 May 22;119(10):6326-6369. doi: 10.1021/acs.chemrev.8b00580. Epub 2019 Feb 4. Chem Rev. 2019. PMID: 30714375 Review.
-
Modeling DNA-Strand Displacement Reactions in the Presence of Base-Pair Mismatches.J Am Chem Soc. 2020 Jul 1;142(26):11451-11463. doi: 10.1021/jacs.0c03105. Epub 2020 Jun 22. J Am Chem Soc. 2020. PMID: 32496760
-
Toehold clipping: A mechanism for remote control of DNA strand displacement.Nucleic Acids Res. 2023 May 8;51(8):4055-4063. doi: 10.1093/nar/gkac1152. Nucleic Acids Res. 2023. PMID: 36477864 Free PMC article.
Cited by
-
NucleoCraft: The Art of Stimuli-Responsive Precision in DNA and RNA Bioengineering.BME Front. 2024 Sep 17;5:0050. doi: 10.34133/bmef.0050. eCollection 2024. BME Front. 2024. PMID: 39290204 Free PMC article. Review.
-
Synthetic RNA biology.RNA Biol. 2024 Jan;21(1):1-2. doi: 10.1080/15476286.2024.2335746. Epub 2024 Apr 14. RNA Biol. 2024. PMID: 38616320 Free PMC article. No abstract available.
-
Induction of bacterial expression at the mRNA level by light.Nucleic Acids Res. 2024 Sep 9;52(16):10017-10028. doi: 10.1093/nar/gkae678. Nucleic Acids Res. 2024. PMID: 39126322 Free PMC article.
References
-
- Simmel FC, Yurke B, Singh HR.. Principles and applications of nucleic acid strand displacement reactions. Chem Rev 2019. May 22;119(10):6326–6369. - PubMed
-
- Lee CS, Davis RW, Davidson N.. A physical study by electron microscopy of the terminally repetitious, circularly permuted DNA from the coliphage particles of Escherichia coli 15. J Mol Biol. 1970;48(1):1–22. - PubMed
-
- Broker TR, Lehman IR. Branched Dna molecules - intermediates in T4 recombination. J Mol Biol. 1971;60(1):131- &. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
