Prior metabolite extraction fully preserves RNAseq quality and enables integrative multi-'omics analysis of the liver metabolic response to viral infection
- PMID: 37095747
- PMCID: PMC10132226
- DOI: 10.1080/15476286.2023.2204586
Prior metabolite extraction fully preserves RNAseq quality and enables integrative multi-'omics analysis of the liver metabolic response to viral infection
Abstract
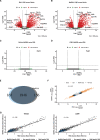
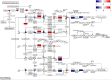
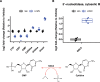
Here, we provide an in-depth analysis of the usefulness of single-sample metabolite/RNA extraction for multi-'omics readout. Using pulverized frozen livers of mice injected with lymphocytic choriomeningitis virus (LCMV) or vehicle (Veh), we isolated RNA prior (RNA) or following metabolite extraction (MetRNA). RNA sequencing (RNAseq) data were evaluated for differential expression analysis and dispersion, and differential metabolite abundance was determined. Both RNA and MetRNA clustered together by principal component analysis, indicating that inter-individual differences were the largest source of variance. Over 85% of LCMV versus Veh differentially expressed genes were shared between extraction methods, with the remaining 15% evenly and randomly divided between groups. Differentially expressed genes unique to the extraction method were attributed to randomness around the 0.05 FDR cut-off and stochastic changes in variance and mean expression. In addition, analysis using the mean absolute difference showed no difference in the dispersion of transcripts between extraction methods. Altogether, our data show that prior metabolite extraction preserves RNAseq data quality, which enables us to confidently perform integrated pathway enrichment analysis on metabolomics and RNAseq data from a single sample. This analysis revealed pyrimidine metabolism as the most LCMV-impacted pathway. Combined analysis of genes and metabolites in the pathway exposed a pattern in the degradation of pyrimidine nucleotides leading to uracil generation. In support of this, uracil was among the most differentially abundant metabolites in serum upon LCMV infection. Our data suggest that hepatic uracil export is a novel phenotypic feature of acute infection and highlight the usefulness of our integrated single-sample multi-'omics approach.
Keywords: RNA; Transcriptomics; integrated omics; mass spectrometry; metabolomics; systems biology.
Conflict of interest statement
RGJ is a scientific advisor for Agios Pharmaceuticals and Servier Pharmaceuticals and is a member of the Scientific Advisory Board of Immunomet Therapeutics.
Figures



References
-
- Inigo M, Deja S, Burgess SC. Ins and outs of the TCA cycle: the central role of anaplerosis. Annu Rev Nutr. 2021;41(1):19–47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
