Statistical learning of protein elastic network from positional covariance matrix
- PMID: 37095762
- PMCID: PMC10121796
- DOI: 10.1016/j.csbj.2023.03.033
Statistical learning of protein elastic network from positional covariance matrix
Abstract
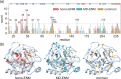
Positional fluctuation and covariance during protein dynamics are key observables for understanding the molecular origin of biological functions. A frequently employed potential energy function for describing protein structural variation at the coarse-gained level is elastic network model (ENM). A long-standing issue in biomolecular simulation is thus the parametrization of ENM spring constants from the components of positional covariance matrix (PCM). Based on sensitivity analysis of PCM, the direct-coupling statistics of each spring, which is a specific combination of position fluctuation and covariance, is found to exhibit prominent signal of parameter dependence. This finding provides the basis for devising the objective function and the scheme of running through the effective one-dimensional optimization of every spring by self-consistent iteration. Formal derivation of the positional covariance statistical learning (PCSL) method also motivates the necessary data regularization for stable calculations. Robust convergence of PCSL is achieved in taking an all-atom molecular dynamics trajectory or an ensemble of homologous structures as input data. The PCSL framework can also be generalized with mixed objective functions to capture specific property such as the residue flexibility profile. Such physical chemistry-based statistical learning thus provides a useful platform for integrating the mechanical information encoded in various experimental or computational data.
Keywords: All-atom molecular dynamics simulation; Elastic network model; Homologous structure; Positional covariance matrix; Serine protease; Statistical learning.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Edge weights in a protein elastic network reorganize collective motions and render long-range sensitivity responses.J Chem Phys. 2022 Jun 28;156(24):245105. doi: 10.1063/5.0095107. J Chem Phys. 2022. PMID: 35778086
-
Estimating the Directional Flexibility of Proteins from Equilibrium Thermal Fluctuations.J Chem Theory Comput. 2021 May 11;17(5):3103-3118. doi: 10.1021/acs.jctc.0c01070. Epub 2021 Apr 5. J Chem Theory Comput. 2021. PMID: 33818072
-
Mechanical couplings of protein backbone and side chains exhibit scale-free network properties and specific hotspots for function.Comput Struct Biotechnol J. 2021 Sep 8;19:5309-5320. doi: 10.1016/j.csbj.2021.09.004. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34765086 Free PMC article.
-
Calculation of Enzyme Fluctuograms from All-Atom Molecular Dynamics Simulation.Methods Enzymol. 2016;578:327-42. doi: 10.1016/bs.mie.2016.05.024. Epub 2016 Jun 20. Methods Enzymol. 2016. PMID: 27497173 Review.
-
Fluctuation matching approach for elastic network model and structure-based model of biomacromolecules.Prog Biophys Mol Biol. 2017 Sep;128:100-112. doi: 10.1016/j.pbiomolbio.2016.12.006. Epub 2016 Dec 30. Prog Biophys Mol Biol. 2017. PMID: 28043838 Review.
Cited by
-
Mechanical codes of chemical-scale specificity in DNA motifs.Chem Sci. 2023 Aug 29;14(37):10155-10166. doi: 10.1039/d3sc01671d. eCollection 2023 Sep 27. Chem Sci. 2023. PMID: 37772098 Free PMC article.
References
LinkOut - more resources
Full Text Sources
