Comparison of alternative splicing (AS) events in adipose tissue of polled dorset versus small tail han sheep
- PMID: 37095997
- PMCID: PMC10121611
- DOI: 10.1016/j.heliyon.2023.e14938
Comparison of alternative splicing (AS) events in adipose tissue of polled dorset versus small tail han sheep
Abstract
Background: During the alternative splicing (AS), the exons of primary transcripts are spliced in various arrangements, resulting in structurally and functionally distinct mRNAs and proteins. This study aimed to examine genes with AS events from Small Tail Han sheep and Dorset sheep to explore the mechanism of adipose developments.
Methods: This study identified the genes with AS events in adipose tissues of two different sheep with next-generation sequencing. In this paper, genes with significantly different AS events were performed gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses.
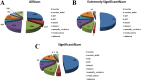
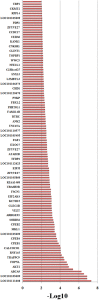
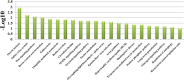
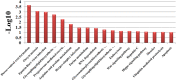
Results: 364 genes with 411 A S events showed significant differences in adipose tissues between the two breeds; 108 genes with 120 A S events were extremely significant differences between the two breeds. We identified several novel genes that are related with adipose growth and development. The results of KEGG and GO analysis indicated that oocyte meiosis, mitogen-activated protein kinase (Wnt), mitogen-activated protein kinase (MAPK) signaling pathway, etc. Were closely related to the adipose tissue developments.
Conclusions: This paper revealed that the genes with AS events are important for adipose tissues in sheep, exploring the mechanisms of AS events associated with adipose tissue developments in sheep of different breeds.
Keywords: Adipose; Alternative splicing; Sheep; Transcriptome; mRNA.
©2023PublishedbyElsevierLtd.
Conflict of interest statement
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Figures

Similar articles
-
Deciphering the role of alternative splicing as a potential regulator in fat-tail development of sheep: a comprehensive RNA-seq based study.Sci Rep. 2024 Jan 29;14(1):2361. doi: 10.1038/s41598-024-52855-1. Sci Rep. 2024. PMID: 38287039 Free PMC article. Review.
-
Comparative analysis of alternative splicing events in skeletal muscle of different sheep.Heliyon. 2023 Nov 8;9(11):e22118. doi: 10.1016/j.heliyon.2023.e22118. eCollection 2023 Nov. Heliyon. 2023. PMID: 38034685 Free PMC article.
-
Ovarian transcriptomic analysis reveals the alternative splicing events associated with fecundity in different sheep breeds.Anim Reprod Sci. 2018 Nov;198:177-183. doi: 10.1016/j.anireprosci.2018.09.017. Epub 2018 Sep 29. Anim Reprod Sci. 2018. PMID: 30318312
-
Transcriptome analysis of adipose tissues from two fat-tailed sheep breeds reveals key genes involved in fat deposition.BMC Genomics. 2018 May 8;19(1):338. doi: 10.1186/s12864-018-4747-1. BMC Genomics. 2018. PMID: 29739312 Free PMC article.
-
Alternative splicing and tissue expression of CIB4 gene in sheep testis.Anim Reprod Sci. 2010 Jul;120(1-4):1-9. doi: 10.1016/j.anireprosci.2010.01.004. Epub 2010 Feb 12. Anim Reprod Sci. 2010. PMID: 20236775 Review.
Cited by
-
A gene expression atlas of Nicotiana tabacum across various tissues at transcript resolution.Front Plant Sci. 2025 Feb 7;16:1500654. doi: 10.3389/fpls.2025.1500654. eCollection 2025. Front Plant Sci. 2025. PMID: 39980486 Free PMC article.
-
Transcriptome-Metabolome Analysis Reveals That Crossbreeding Improves Meat Quality in Hu Sheep and Their F1-Generation Sheep.Foods. 2025 Apr 17;14(8):1384. doi: 10.3390/foods14081384. Foods. 2025. PMID: 40282783 Free PMC article.
-
Deciphering the role of alternative splicing as a potential regulator in fat-tail development of sheep: a comprehensive RNA-seq based study.Sci Rep. 2024 Jan 29;14(1):2361. doi: 10.1038/s41598-024-52855-1. Sci Rep. 2024. PMID: 38287039 Free PMC article. Review.
-
Comparative analysis of alternative splicing events in skeletal muscle of different sheep.Heliyon. 2023 Nov 8;9(11):e22118. doi: 10.1016/j.heliyon.2023.e22118. eCollection 2023 Nov. Heliyon. 2023. PMID: 38034685 Free PMC article.
References
-
- Atti N., Bocquier F., Khaldi G. Performance of the fat-tailed Barbarine sheep in its environment: adaptive capacity to alternation of underfeeding and re-feeding periods. Anim. Res. 2004;53:165–176.
-
- Miao X.Y., Luo Q.M., Qin X.Y. Genome-wide transcriptome analysis of mRNAs and microRNAs in Dorset and Small Tail Han sheep to explore the regulation of fecundity. Mol. Cell. Endocrinol. 2015;40:232–242. - PubMed
-
- Miao X.Y., Luo Q.M., Qin X.Y., Guo Y.Y., Zhao H.J. Genome-wide mRNA-seq profiling reveals predominant down-regulation of lipid metabolic processes in adipose tissues of Small Tail Han than Dorset sheep. Biochem. Biophys. Res. Commun. 2015;467:413–420. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

