Gram-positive anaerobic cocci guard skin homeostasis by regulating host-defense mechanisms
- PMID: 37096035
- PMCID: PMC10122035
- DOI: 10.1016/j.isci.2023.106483
Gram-positive anaerobic cocci guard skin homeostasis by regulating host-defense mechanisms
Abstract
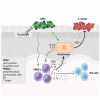
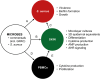
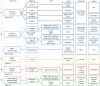
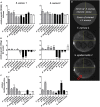
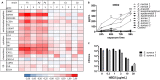
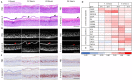
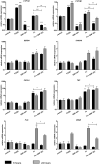
In atopic dermatitis (AD), chronic skin inflammation is associated with skin barrier defects and skin microbiome dysbiosis including a lower abundance of Gram-positive anaerobic cocci (GPACs). We here report that, through secreted soluble factors, GPAC rapidly and directly induced epidermal host-defense molecules in cultured human keratinocytes and indirectly via immune-cell activation and cytokines derived thereof. Host-derived antimicrobial peptides known to limit the growth of Staphylococcus aureus-a skin pathogen involved in AD pathology-were strongly upregulated by GPAC-induced signaling through aryl hydrocarbon receptor (AHR)-independent mechanisms, with a concomitant AHR-dependent induction of epidermal differentiation genes and control of pro-inflammatory gene expression in organotypic human epidermis. By these modes of operandi, GPAC may act as an "alarm signal" and protect the skin from pathogenic colonization and infection in the event of skin barrier disruption. Fostering growth or survival of GPAC may be starting point for microbiome-targeted therapeutics in AD.
Keywords: Dermatology; Microbiology; microbiome.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Altunbulakli C., Reiger M., Neumann A.U., Garzorz-Stark N., Fleming M., Huelpuesch C., Castro-Giner F., Eyerich K., Akdis C.A., Traidl-Hoffmann C. Relations between epidermal barrier dysregulation and Staphylococcus species–dominated microbiome dysbiosis in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2018;142:1643–1647.e12. doi: 10.1016/j.jaci.2018.07.005. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

