Analysing transcriptomic signatures and identifying potential genes for the protective effect of inactivated COVID-19 vaccines
- PMID: 37096063
- PMCID: PMC10122457
- DOI: 10.7717/peerj.15155
Analysing transcriptomic signatures and identifying potential genes for the protective effect of inactivated COVID-19 vaccines
Abstract
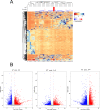
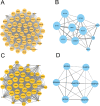
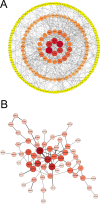
Inactivated vaccines are one of the most effective strategies for controlling the coronavirus disease 2019 (COVID-19) pandemic. However, the response genes for the protective effect of inactivated vaccines are still unclear. Herein, we analysed the neutralization antibody responses elicited by vaccine serum and carried out transcriptome sequencing of RNAs isolated from the PBMCs of 29 medical staff receiving two doses of the CoronaVac vaccine. The results showed that SARS-CoV-2 neutralization antibody titers varied considerably among individuals, and revealed that many innate immune pathways were activated after vaccination. Furthermore, the blue module revealed that NRAS, YWHAB, SMARCA5, PPP1CC and CDC5L may be correlated with the protective effect of the inactivated vaccine. Additionally, MAPK1, CDC42, PPP2CA, EP300, YWHAZ and NRAS were demonstrated as the hub genes having a significant association with vaccines. These findings provide a basis for understanding the molecular mechanism of the host immune response induced by inactivated vaccines.
Keywords: Inactivated vaccine; Neutralization antibody; RNA-seq; SARS-CoV-2; Transcriptome profile.
©2023 Chen et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


References
-
- Abu-Raddad LJ, Chemaitelly H, Ayoub HH, Coyle P, Malek JA, Ahmed AA, Mohamoud YA, Younuskunju S, Tang P, Al Kanaani Z, Al Kuwari E, Butt AA, Jeremijenko A, Kaleeckal AH, Latif AN, Shaik RM, Abdul Rahim HF, Nasrallah GK, Yassine HM, Al Kuwari MG, Al Romaihi HE, Al-Thani MH, Al Khal A, Bertollini R. Introduction and expansion of the SARS-CoV-2 B.1.1.7 variant and reinfections in Qatar: a nationally representative cohort study. PLOS Medicine. 2021;18:e1003879. doi: 10.1371/journal.pmed.1003879. - DOI - PMC - PubMed
-
- Alcorn JF, Avula R, Chakka AB, Schwarzmann WE, Nowalk MP, Lin CJ, Ortiz MA, Horne WT, Chandran UR, Nagg JP, Zimmerman RK, Cole KS, Moehling KK, Martin JM. Differential gene expression in peripheral blood mononuclear cells from children immunized with inactivated influenza vaccine. Human Vaccines & Immunotherapeutics. 2020;16:1782–1790. doi: 10.1080/21645515.2020.1711677. - DOI - PMC - PubMed
-
- Ali K, Berman G, Zhou H, Deng W, Faughnan V, Coronado-Voges M, Ding B, Dooley J, Girard B, Hillebrand W, Pajon R, Miller JM, Leav B, McPhee R. Evaluation of mRNA-1273 SARS-CoV-2 Vaccine in Adolescents. The New England Journal of Medicine. 2021;385:2241–2251. doi: 10.1056/NEJMoa2109522. - DOI - PMC - PubMed
-
- Arunachalam PS, Scott MKD, Hagan T, Li C, Feng Y, Wimmers F, Grigoryan L, Trisal M, Edara VV, Lai L, Chang SE, Feng A, Dhingra S, Shah M, Lee AS, Chinthrajah S, Sindher SB, Mallajosyula V, Gao F, Sigal N, Kowli S, Gupta S, Pellegrini K, Tharp G, Maysel-Auslender S, Hamilton S, Aoued H, Hrusovsky K, Roskey M, Bosinger SE, Maecker HT, Boyd SD, Davis MM, Utz PJ, Suthar MS, Khatri P, Nadeau KC, Pulendran B. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature. 2021;596:410–416. doi: 10.1038/s41586-021-03791-x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

