Examining the Early Distribution of the Artemisinin-Resistant Plasmodium falciparum kelch13 R561H Mutation in Areas of Higher Transmission in Rwanda
- PMID: 37096145
- PMCID: PMC10122489
- DOI: 10.1093/ofid/ofad149
Examining the Early Distribution of the Artemisinin-Resistant Plasmodium falciparum kelch13 R561H Mutation in Areas of Higher Transmission in Rwanda
Abstract
Background: Artemisinin resistance mutations in Plasmodium falciparum kelch13 (Pfk13) have begun to emerge in Africa, with Pfk13-R561H being the first reported in Rwanda in 2014, but limited sampling left questions about its early distribution and origin.
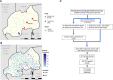
Methods: We genotyped P. falciparum positive dried blood spot (DBS) samples from a nationally representative 2014-2015 Rwanda Demographic Health Surveys (DHS) HIV study. DBS were subsampled from DHS sampling clusters with >15% P. falciparum prevalence, as determined by rapid testing or microscopy done during the DHS study (n clusters = 67, n samples = 1873).
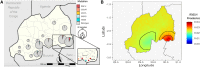
Results: We detected 476 parasitemias among 1873 residual blood spots from a 2014-2015 Rwanda Demographic Health Survey. We sequenced 351 samples: 341/351 were wild-type (97.03% weighted), and 4 samples (1.34% weighted) harbored R561H that were significantly spatially clustered. Other nonsynonymous mutations found were V555A (3), C532W (1), and G533A (1).
Conclusions: Our study better defines the early distribution of R561H in Rwanda. Previous studies only observed the mutation in Masaka as of 2014, but our study indicates its presence in higher-transmission regions in the southeast of the country at that time.
Keywords: antimalarial resistance; artemisinin; drug resistance; k13; kelch13; r561H.
© The Author(s) 2023. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest. All authors: no reported conflicts.
Figures
Similar articles
-
A review of the frequencies of Plasmodium falciparum Kelch 13 artemisinin resistance mutations in Africa.Int J Parasitol Drugs Drug Resist. 2021 Aug;16:155-161. doi: 10.1016/j.ijpddr.2021.06.001. Epub 2021 Jun 10. Int J Parasitol Drugs Drug Resist. 2021. PMID: 34146993 Free PMC article. Review.
-
High Prevalence of Plasmodium falciparum K13 Mutations in Rwanda Is Associated With Slow Parasite Clearance After Treatment With Artemether-Lumefantrine.J Infect Dis. 2022 Apr 19;225(8):1411-1414. doi: 10.1093/infdis/jiab352. J Infect Dis. 2022. PMID: 34216470 Free PMC article. Clinical Trial.
-
Emergence and Spread of kelch13 Mutations Associated with Artemisinin Resistance in Plasmodium falciparum Parasites in 12 Thai Provinces from 2007 to 2016.Antimicrob Agents Chemother. 2018 Mar 27;62(4):e02141-17. doi: 10.1128/AAC.02141-17. Print 2018 Apr. Antimicrob Agents Chemother. 2018. PMID: 29378723 Free PMC article.
-
Novel Plasmodium falciparum k13 gene polymorphisms from Kisii County, Kenya during an era of artemisinin-based combination therapy deployment.Malar J. 2023 Mar 9;22(1):87. doi: 10.1186/s12936-023-04517-2. Malar J. 2023. PMID: 36894982 Free PMC article.
-
Artemisinin and multidrug-resistant Plasmodium falciparum - a threat for malaria control and elimination.Curr Opin Infect Dis. 2021 Oct 1;34(5):432-439. doi: 10.1097/QCO.0000000000000766. Curr Opin Infect Dis. 2021. PMID: 34267045 Free PMC article. Review.
Cited by
-
Temporal genomics in Southern Zambia shows rising prevalence of Plasmodium falciparum mutations linked to delayed clearance after artemisinin-lumefantrine treatment.Sci Rep. 2024 Nov 5;14(1):26789. doi: 10.1038/s41598-024-76442-6. Sci Rep. 2024. PMID: 39500918 Free PMC article.
-
Anti-malarial resistance in Mozambique: Absence of Plasmodium falciparum Kelch 13 (K13) propeller domain polymorphisms associated with resistance to artemisinins.Malar J. 2023 May 19;22(1):160. doi: 10.1186/s12936-023-04589-0. Malar J. 2023. PMID: 37208708 Free PMC article.
-
High Frequency of Artemisinin Partial Resistance Mutations in the Great Lakes Region Revealed Through Rapid Pooled Deep Sequencing.J Infect Dis. 2025 Feb 4;231(1):269-280. doi: 10.1093/infdis/jiae475. J Infect Dis. 2025. PMID: 39367758
-
Understanding Malaria Treatment Adherence in Rwanda: Implications for Artemisinin Resistance.medRxiv [Preprint]. 2025 Jul 23:2025.07.22.25331991. doi: 10.1101/2025.07.22.25331991. medRxiv. 2025. PMID: 40778167 Free PMC article. Preprint.
-
Prevalence of Asymptomatic non-Falciparum and Falciparum Malaria in the 2014-15 Rwanda Demographic Health Survey.medRxiv [Preprint]. 2024 Dec 12:2024.01.09.24301054. doi: 10.1101/2024.01.09.24301054. medRxiv. 2024. PMID: 38260604 Free PMC article. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

