DNA Damage-Induced, S-Phase Specific Phosphorylation of Orc6 is Critical for the Maintenance of Genome Stability
- PMID: 37096556
- PMCID: PMC10153009
- DOI: 10.1080/10985549.2023.2196204
DNA Damage-Induced, S-Phase Specific Phosphorylation of Orc6 is Critical for the Maintenance of Genome Stability
Abstract
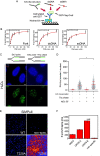
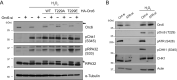
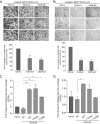
The smallest subunit of the human Origin Recognition Complex, hOrc6, is required for DNA replication progression and plays an important role in mismatch repair (MMR) during S-phase. However, the molecular details of how hOrc6 regulates DNA replication and DNA damage response remain to be elucidated. Orc6 levels are elevated upon specific types of genotoxic stress, and it is phosphorylated at Thr229, predominantly during S-phase, in response to oxidative stress. Many repair pathways, including MMR, mediate oxidative DNA damage repair. Defects in MMR are linked to Lynch syndrome, predisposing patients to many cancers, including colorectal cancer. Orc6 levels are known to be elevated in colorectal cancers. Interestingly, tumor cells show reduced hOrc6-Thr229 phosphorylation compared to adjacent normal mucosa. Further, elevated expression of wild-type and the phospho-dead forms of Orc6 results in increased tumorigenicity, implying that in the absence of this "checkpoint" signal, cells proliferate unabated. Based on these results, we propose that DNA-damage-induced hOrc6-pThr229 phosphorylation during S-phase facilitates ATR signaling in the S-phase, halts fork progression, and enables assembly of repair factors to mediate efficient repair to prevent tumorigenesis. Our study provides novel insights into how hOrc6 regulates genome stability.
Keywords: DNA damage; Orc6; phosphorylation; replication.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
