Genome-wide identification of the glutamate receptor-like gene family in Vanilla planifolia and their response to Fusarium oxysporum infection
- PMID: 37096589
- PMCID: PMC10132242
- DOI: 10.1080/15592324.2023.2204654
Genome-wide identification of the glutamate receptor-like gene family in Vanilla planifolia and their response to Fusarium oxysporum infection
Abstract
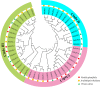
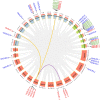
Glutamate receptor-like genes (GLRs) are essential for plant growth and development and for coping with environmental (biological and non-biological) stresses. In this study, 13 GLR members were identified in the Vanilla planifolia genome and attributed to two subgroups (Clade I and Clade III) based on their physical relationships. Cis-acting element analysis and Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations indicated the GLR gene regulation's complexity and their functional diversity. Expression analysis revealed a relatively higher and more general expression pattern of Clade III members compared to the Clade I subgroup in tissues. Most GLRs showed significant differences in expression during Fusarium oxysporum infection. This suggested that GLRs play a critical role in the response of V. planifolia to pathogenic infection. These results provide helpful information for further functional research and crop improvement of VpGLRs.
Keywords: Fusarium oxysporum; Vanilla planifolia; biotic stress; expression pattern; glutamate receptor-like genes.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures





References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
