Machine learning for RNA 2D structure prediction benchmarked on experimental data
- PMID: 37096592
- PMCID: PMC10199776
- DOI: 10.1093/bib/bbad153
Machine learning for RNA 2D structure prediction benchmarked on experimental data
Abstract
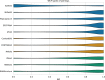
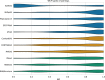
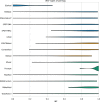
Since the 1980s, dozens of computational methods have addressed the problem of predicting RNA secondary structure. Among them are those that follow standard optimization approaches and, more recently, machine learning (ML) algorithms. The former were repeatedly benchmarked on various datasets. The latter, on the other hand, have not yet undergone extensive analysis that could suggest to the user which algorithm best fits the problem to be solved. In this review, we compare 15 methods that predict the secondary structure of RNA, of which 6 are based on deep learning (DL), 3 on shallow learning (SL) and 6 control methods on non-ML approaches. We discuss the ML strategies implemented and perform three experiments in which we evaluate the prediction of (I) representatives of the RNA equivalence classes, (II) selected Rfam sequences and (III) RNAs from new Rfam families. We show that DL-based algorithms (such as SPOT-RNA and UFold) can outperform SL and traditional methods if the data distribution is similar in the training and testing set. However, when predicting 2D structures for new RNA families, the advantage of DL is no longer clear, and its performance is inferior or equal to that of SL and non-ML methods.
Keywords: RNA 2D structure prediction; algorithm benchmarking; deep learning; machine learning.
© The Author(s) 2023. Published by Oxford University Press.
Figures
Similar articles
-
UFold: fast and accurate RNA secondary structure prediction with deep learning.Nucleic Acids Res. 2022 Feb 22;50(3):e14. doi: 10.1093/nar/gkab1074. Nucleic Acids Res. 2022. PMID: 34792173 Free PMC article.
-
MSFF-CDCGAN: A novel method to predict RNA secondary structure based on Generative Adversarial Network.Methods. 2022 Aug;204:368-375. doi: 10.1016/j.ymeth.2022.04.004. Epub 2022 Apr 28. Methods. 2022. PMID: 35490852
-
Machine Learning Analysis of RNA-seq Data for Diagnostic and Prognostic Prediction of Colon Cancer.Sensors (Basel). 2023 Mar 13;23(6):3080. doi: 10.3390/s23063080. Sensors (Basel). 2023. PMID: 36991790 Free PMC article.
-
Review of machine learning methods for RNA secondary structure prediction.PLoS Comput Biol. 2021 Aug 26;17(8):e1009291. doi: 10.1371/journal.pcbi.1009291. eCollection 2021 Aug. PLoS Comput Biol. 2021. PMID: 34437528 Free PMC article. Review.
-
Secondary structure prediction of long noncoding RNA: review and experimental comparison of existing approaches.Brief Bioinform. 2022 Jul 18;23(4):bbac205. doi: 10.1093/bib/bbac205. Brief Bioinform. 2022. PMID: 35692094 Review.
Cited by
-
RNA secondary structure prediction by conducting multi-class classifications.Comput Struct Biotechnol J. 2025 Apr 4;27:1449-1459. doi: 10.1016/j.csbj.2025.04.001. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40256169 Free PMC article.
-
Systematic benchmarking of deep-learning methods for tertiary RNA structure prediction.PLoS Comput Biol. 2024 Dec 30;20(12):e1012715. doi: 10.1371/journal.pcbi.1012715. eCollection 2024 Dec. PLoS Comput Biol. 2024. PMID: 39775239 Free PMC article.
-
Analysis of natural structures and chemical mapping data reveals local stability compensation in RNA.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf565. doi: 10.1093/nar/gkaf565. Nucleic Acids Res. 2025. PMID: 40568944 Free PMC article.
-
Comprehensive datasets for RNA design, machine learning, and beyond.Sci Rep. 2025 Jul 1;15(1):21417. doi: 10.1038/s41598-025-07041-2. Sci Rep. 2025. PMID: 40594473 Free PMC article.
-
Comparative analysis of RNA 3D structure prediction methods: towards enhanced modeling of RNA-ligand interactions.Nucleic Acids Res. 2024 Jul 22;52(13):7465-7486. doi: 10.1093/nar/gkae541. Nucleic Acids Res. 2024. PMID: 38917327 Free PMC article.
References
-
- Mortimer SA, Kidwell MA, Doudna JA. Insights into RNA structure and function from genome-wide studies. Nat Rev Genet 2014;15(7):469–79. - PubMed
-
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature 2004;431:343–9. - PubMed
-
- Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell 2008;29(1):1–7. - PubMed

