First-Day-of-Life Rectal Swabs Fail To Represent Meconial Microbiota Composition and Underestimate the Presence of Antibiotic Resistance Genes
- PMID: 37097170
- PMCID: PMC10269712
- DOI: 10.1128/spectrum.05254-22
First-Day-of-Life Rectal Swabs Fail To Represent Meconial Microbiota Composition and Underestimate the Presence of Antibiotic Resistance Genes
Abstract
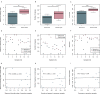
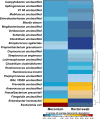
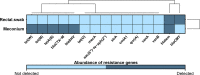
The human gut microbiome plays a vital role in health and disease. In particular, the first days of life provide a unique window of opportunity for development and establishment of microbial community. Currently, stool samples are known to be the most widely used sampling approach for studying the gut microbiome. However, complicated sample acquisition at certain time points, challenges in transportation, and patient discomfort underline the need for development of alternative sampling approaches. One of the alternatives is rectal swabs, shown to be a reliable proxy for gut microbiome analysis when obtained from adults. Here, we compare the usability of rectal swabs and meconium paired samples collected from infants on the first days of life. Our results indicate that the two sampling approaches display significantly distinct patterns in microbial composition and alpha and beta diversity as well as detection of resistance genes. Moreover, the dissimilarity between the two collection methods was greater than the interindividual variation. Therefore, we conclude that rectal swabs are not a reliable proxy compared to stool samples for gut microbiome analysis when collected on the first days of a newborn's life. IMPORTANCE Currently, there are numerous suggestions on how to ease the notoriously complex and error-prone methodological setups to study the gut microbiota of newborns during the first days of life. Especially, meconium samples are regularly failing to yield meaningful data output and therefore have been suggested to be replaced by rectal swabs as done in adults as well. We find this development toward a simplified method to be producing dramatically erroneous results, skewing data interpretation away from the real aspects to be considered for neonatal health during the first days of life. We have put together our knowledge on this critical aspect with careful consideration and identified the failure of rectal swabs to be a replacement for sampling of meconium in term-born newborns.
Keywords: antibiotic resistance; gut microbiota; human microbiota; meconium; microbiome; newborns; rectal swabs; resistance genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Wong WS, Sabu P, Deopujari V, Levy S, Shah AA, Clemency N, Provenzano M, Saadoon R, Munagala A, Baker R, Baveja R, Mueller NT, Dominguez-Bello MG, Huddleston K, Niederhuber JE, Hourigan SK. 2020. Prenatal and peripartum exposure to antibiotics and cesarean section delivery are associated with differences in diversity and composition of the infant meconium microbiome. Microorganisms 8:179. doi:10.3390/microorganisms8020179. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

