TRAF3-EWSR1 signaling axis acts as a checkpoint on germinal center responses
- PMID: 37097293
- PMCID: PMC10130905
- DOI: 10.1084/jem.20221483
TRAF3-EWSR1 signaling axis acts as a checkpoint on germinal center responses
Erratum in
-
Correction: TRAF3-EWSR1 signaling axis acts as a checkpoint on germinal center responses.J Exp Med. 2023 Aug 7;220(8):e2022148307122023c. doi: 10.1084/jem.2022148307122023c. Epub 2023 Jul 18. J Exp Med. 2023. PMID: 37462917 Free PMC article. No abstract available.
Abstract
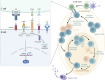
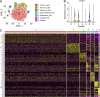
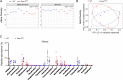
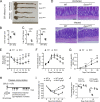
The formation of germinal centers (GCs) is crucial for humoral immunity and vaccine efficacy. Constant stimulation through microbiota drives the formation of constitutive GCs in Peyer's patches (PPs), which generate B cells that produce antibodies against gut antigens derived from commensal bacteria and infectious pathogens. However, the molecular mechanism that regulates this persistent process is poorly understood. We report that Ewing Sarcoma Breakpoint Region 1 (EWSR1) is a brake to constitutive GC generation and immunoglobulin G (IgG) production in PPs, vaccination-induced GC formation, and IgG responses. Mechanistically, EWSR1 suppresses Bcl6 upregulation after antigen encounter, thereby negatively regulating induced GC B cell generation and IgG production. We further showed that tumor necrosis factor receptor-associated factor (TRAF) 3 serves as a negative regulator of EWSR1. These results established that the TRAF3-EWSR1 signaling axis acts as a checkpoint for Bcl6 expression and GC responses, indicating that this axis is a therapeutic target to tune GC responses and humoral immunity in infectious diseases.
© 2023 Li et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures









Similar articles
-
Antigen receptor signaling and cell death resistance controls intestinal humoral response zonation.Immunity. 2023 Oct 10;56(10):2373-2387.e8. doi: 10.1016/j.immuni.2023.08.018. Epub 2023 Sep 14. Immunity. 2023. PMID: 37714151 Free PMC article.
-
Affinity-based clonal selection in Peyer's patches.Curr Opin Immunol. 2022 Feb;74:100-105. doi: 10.1016/j.coi.2021.11.002. Epub 2021 Nov 27. Curr Opin Immunol. 2022. PMID: 34847473 Review.
-
Dietary Antigens Induce Germinal Center Responses in Peyer's Patches and Antigen-Specific IgA Production.Front Immunol. 2019 Oct 15;10:2432. doi: 10.3389/fimmu.2019.02432. eCollection 2019. Front Immunol. 2019. PMID: 31681315 Free PMC article.
-
BCR affinity differentially regulates colonization of the subepithelial dome and infiltration into germinal centers within Peyer's patches.Nat Immunol. 2019 Apr;20(4):482-492. doi: 10.1038/s41590-019-0325-1. Epub 2019 Mar 4. Nat Immunol. 2019. PMID: 30833793
-
Impaired selection of IgA and intestinal dysbiosis associated with PD-1-deficiency.Gut Microbes. 2013 Mar-Apr;4(2):165-71. doi: 10.4161/gmic.23595. Epub 2013 Jan 18. Gut Microbes. 2013. PMID: 23333864 Free PMC article. Review.
Cited by
-
PGRN Inhibits Early B-cell Activation and IgE Production Through the IFITM3-STAT1 Signaling Pathway in Asthma.Adv Sci (Weinh). 2024 Dec;11(45):e2403939. doi: 10.1002/advs.202403939. Epub 2024 Oct 16. Adv Sci (Weinh). 2024. PMID: 39412083 Free PMC article.
References
-
- Araya, N., Hirota K., Shimamoto Y., Miyagishi M., Yoshida E., Ishida J., Kaneko S., Kaneko M., Nakajima T., and Fukamizu A.. 2003. Cooperative interaction of EWS with CREB-binding protein selectively activates hepatocyte nuclear factor 4-mediated transcription. J. Biol. Chem. 278:5427–5432. 10.1074/jbc.M210234200 - DOI - PubMed
-
- Aurias, A., Rimbaut C., Buffe D., Dubousset J., and Mazabraud A.. 1983. Translocation of chromosome 22 in Ewing’s sarcoma. C. R. Seances Acad. Sci. III. 296:1105–1107. - PubMed
-
- Beccaria, C.G., Amezcua Vesely M.C., Fiocca Vernengo F., Gehrau R.C., Ramello M.C., Tosello Boari J., Gorosito Serrán M., Mucci J., Piaggio E., Campetella O., et al. . 2018. Galectin-3 deficiency drives lupus-like disease by promoting spontaneous germinal centers formation via IFN-γ. Nat. Commun. 9:1628. 10.1038/s41467-018-04063-5 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

