Depleting CD103+ resident memory T cells in vivo reveals immunostimulatory functions in oral mucosa
- PMID: 37097449
- PMCID: PMC10130744
- DOI: 10.1084/jem.20221853
Depleting CD103+ resident memory T cells in vivo reveals immunostimulatory functions in oral mucosa
Abstract
The oral mucosa is a frontline for microbial exposure and juxtaposes several unique tissues and mechanical structures. Based on parabiotic surgery of mice receiving systemic viral infections or co-housing with microbially diverse pet shop mice, we report that the oral mucosa harbors CD8+ CD103+ resident memory T cells (TRM), which locally survey tissues without recirculating. Oral antigen re-encounter during the effector phase of immune responses potentiated TRM establishment within tongue, gums, palate, and cheek. Upon reactivation, oral TRM triggered changes in somatosensory and innate immune gene expression. We developed in vivo methods for depleting CD103+ TRM while sparing CD103neg TRM and recirculating cells. This revealed that CD103+ TRM were responsible for inducing local gene expression changes. Oral TRM putatively protected against local viral infection. This study provides methods for generating, assessing, and in vivo depleting oral TRM, documents their distribution throughout the oral mucosa, and provides evidence that TRM confer protection and trigger responses in oral physiology and innate immunity.
© 2023 Stolley et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures



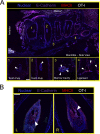
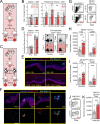
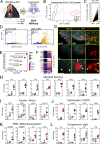
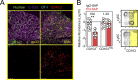


Comment in
-
Guardians of the oral barriers.J Exp Med. 2023 Jul 3;220(7):e20230550. doi: 10.1084/jem.20230550. Epub 2023 May 15. J Exp Med. 2023. PMID: 37184540 Free PMC article.
References
-
- Baller, J., Kono T., Herman A., and Zhang Y.. 2019. CHURP: A lightweight cli framework to enable novice users to analyze sequencing datasets in parallel. In. Proceedings of the Practice and Experience in Advanced Research Computing on Rise of theMachines (Learning), PEARC ’19. Association for Computing Machinery, New York, NY, USA. 1–5. 10.1145/3332186.3333156 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

